Abstract
In the course of discovering new metabolites from co-culture of Penicillium pinophilum FKI-5653 and Trichoderma harzianum FKI-5655, a new compound, designated secopenicillide C, and four known compounds, penicillide, MC-141, pestalasin A and stromemycin were isolated. The production of these compounds, except pestalasin A, was enhanced in co-culture to be two to six times higher than in pure culture of Penicillium pinophilum FKI-5653, which was the producer of these compounds.
Similar content being viewed by others
Introduction
Natural products are useful to develop drugs and other chemical agents. However, the rate of known compounds re-isolated from natural sources were increasing during this decade.1 So, more efficient methods to discover new natural compounds is needed. Meanwhile, in the modern microbiology, pure culture is the basic method since Koch's law in 1884. However, analyses of microbial whole genome sequences indicate that microbes encode the genetic information for the biosynthesis of a number of compounds that are not observed when cultured under standard conditions.2 To overcome this problem, co-culture method draws attention to produce novel natural product and increase the productivity of known compounds. Until now, some examples, such as Gram-positive bacteria and Gram-positive bacteria,3, 4 Gram-positive bacteria and fungus,5, 6 Gram-negative bacteria and fungus,7, 8, 9 and fungus and fungus,10, 11, 12, 13 were reported. In this background, we started to apply this co-culture method to discover new compounds. We report here the fermentation, isolation and structure elucidation of compounds obtained from co-culture broth of two fungal strains, Penicillium pinophilum FKI-5653 and Trichoderma harzianum FKI-5655.
Materials and Methods
General
NMR spectra were measured on a Varian XL-400 spectrometer with 1H NMR at 400 MHz and 13C NMR at 100 MHz (Agilent Technologies, Santa Clara, CA, USA). The chemical shifts are expressed in p.p.m. and are referenced to the solvent CD3OD (3.30 p.p.m.) or (CD3)2SO (2.50 p.p.m.) in the 1H NMR spectra, and referenced to the solvent CD3OD (49.0 p.p.m.) or (CD3)2SO (39.5 p.p.m.) in the 13C NMR spectra. FAB–MS and ESI–MS spectra were measured on a JEOL JMS AX-505 HA mass spectrometer and a JEOL AccuTOF apparatus (JEOL, Akishima, Tokyo, Japan). IR spectra (KBr) were taken on a Horiba FT-210 Fourier transform IR spectrometer (Horiba, Kyoto, Japan). UV spectra were measured with a Hitachi U-2810 spectrophotometer (Hitachi, Tokyo, Japan). The optical rotation was measured with a JEOL DIP-370 polarimeter.
Taxonomic studies of the strain FKI-5653 and FKI-5655
Fungal strains FKI-5653 and FKI-5655 were isolated from the soil collected in Hachijo Island, Tokyo, Japan. For the determination of the morphological characteristics of FKI-5653 using the methodology of Pitt,14 the isolate was inoculated as three-point cultures on Czapek yeast extract agar (CYA), malt extract agar (MEA) and 25% glycerol nitrate agar, and grown for 7 days at 25 °C (also at 5 and 37 °C on CYA) in the dark. Although, for the determination of the morphological characteristics of FKI-5655, the isolate was inoculated as one-point cultures on potato dextrose agar (PDA), corn meal dextrose agar (CMD) and synthetic low-nutrient agar (SNA), and grown for 7 days at 25 °C (also at 5 and 37 °C on PDA) in the dark.
Color Harmony Manual, 4th Edition (Container Corporation of America, Chicago)15 was used to determine color names and hue numbers.
For the determination of micro-morphological characteristics, microscopic slides were prepared using colonies from MEA (for FKI-5653) or synthetic low-nutrient agar (for FKI-5655). The slides were examined with a Vanox-S AH-2 microscope (Olympus, Tokyo, Japan), and digital photomicrographs were taken with a DP25 digital camera (Olympus). For scanning electron microscopy of the conidia and conidiophores, agar blocks (5 mm2) were cut from a 7-day-old culture of the strains growing on MEA or SNA. The agar blocks were fixed with osmium tetroxide (TAAB, Berks, UK), air-dried and sputter-coated with gold using a JFC-1200 Fine Coater (JEOL). The samples were observed under a JSM-5600 scanning electron microscope (JEOL).
Genomic DNA of the fungal strains FKI-5653 and FKI-5655 were isolated using the PrepMan Ultra Sample Preparation Reagent (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. Amplification of the rDNA internal transcribed spacer (rDNA ITS) including the 5.8S rDNA was performed using primers ITS1 and ITS4.16 Amplifications were performed in a PCR (Verity 96-well thermal cycler; Applied Biosystems), and the PCR products were purified using a QIAquick, PCR DNA Purification kit protocol (Qiagen, Valencia, CA, USA). The PCR products were sequenced directly in both directions using primers ITS1, ITS2, ITS3 and ITS4, using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequencing products were purified by ethanol/EDTA precipitation, and samples were analyzed on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). Contigs were assembled using the forward and reverse sequences with the SeqMan and SeqBuilder programs from the Lasergene 8 package (DNAStar, Madison, WI, USA). The ITS sequence of the strains were deposited at the DNA Data Bank of Japan (DDBJ) with accession number AB606412 for FKI-5653 and AB606413 for FKI-5655.
Fermentation
The strains FKI-5653 and FKI-5655 were each grown and maintained on an agar slant consisting of 0.1%. glycerol, 0.08% KH2PO4, 0.02% K2HPO4, 0.02% MgSO4·7H2O, 0.02% KCl, 0.2% NaNO3, 0.02% yeast extract and 1.5% agar (adjusted to pH 6.0 before sterilization). A loopful of spores of each strain was inoculated into 100 ml of the seed medium consisting of 2.0% glucose, 0.5% polypeptone (Nihon Pharmaceutical, Tokyo, Japan), 0.2% yeast extract, 0.2% KH2PO4, 0.05% MgSO4·7H2O and 0.1% agar (adjusted to pH 6.0 before sterilization) in a 500-ml Erlenmeyer flask. The flask was incubated on a rotary shaker (210 r.p.m.) at 27 °C for 3 days. For the co-culture, a 1-ml portion of each seed culture of FKI-5653 and FKI-5655 was transferred to 50 or 80 Erlenmeyer flasks of 500 ml, each containing 100 ml of production medium (3.0% brown rice powder, 0.5% glycerol, 0.2% yeast extract, 0.2% potato dextrose broth, 0.2% CH3COONH4, 0.3% NaNO3, 0.05% KCl, 0.05% MgSO4·7H2O, 0.001% FeSO4·7H2O, 0.001% ZnSO4·7H2O and 0.001% CuSO4·5H2O (adjusted to pH 6.0 before sterilization)), and the fermentation on a rotary shaker (210 r.p.m.) was carried out at 27 °C for 6 days. For the pure culture, a 1-ml portion of each seed culture of FKI-5653 or FKI-5565 was transferred to three 500-ml Erlenmeyer flasks containing the production medium, and fermentation on a rotary shaker (210 r.p.m.) was carried out at 27 °C for 6 days, respectively.
Analysis of metabolites by HPLC
The production of metabolites was measured by analytical HPLC under the following conditions: column, Symmetry C18 (2.1φ × 150 mm, Waters, Milford, MA, USA); UV detection, 210 nm; flow rate, 0.2 ml min−1; mobile phase, acetonitrile water with 0.05% phosphoric acid, 30–70% (20 min) for the detection of pestalasin A, MC-141, penicillide and secopenicillide C with retention times of 6.24, 11.2, 14.5 and 18.1 min, respectively, or 5–100% (20 min) for the detection of stromemycin with retention time of 17.9 min.
Results
Taxonomy of strain FKI-5653
Colonies on CYA were 35–36 mm in diameter after 7 days at 25 °C (Figure 1a), floccose, plane, with light yellow (1 ga) aerial mycelium, covered with olive gray (1 ig) conidia, exudate lacking, the reverse were maple (4 le), the margin entire, soluble pigment not produced. Colonies on MEA were 37–38 mm in diameter after 7 days at 25 °C (Figure 1b), less dense than on CYA, colliculose, the surface with light citron yellow (1 hb) floccose aerial mycelium, exudate lacking, the reverse were maize (2 hb), the margin entire, soluble pigment not produced. Colonies on glycerol nitrate agar were 6–7 mm in diameter after 7 days at 25 °C, plane, the surface with light ivory (2 ca) floccose aerial mycelium, exudate lacking, the reverse were light ivory (2 ca), the margin entire, soluble pigment not produced. Colonies on CYA were 35–36 mm in diameter after 7 days at 37 °C, veltinous, plane, radially sulcate, with pastel yellow (1 1/2 fb) aerial mycelium, covered with olive gray (1 1/2 ig) conidia, exudate lacking, the reverse were yellow maple (3 le), the margin entire, soluble pigment not produced. The culture on CYA at 5 °C showed no growth.
Conidiophores on MEA were borne on a basal felt or directly from the agar, stripes were simple or rarely branched, 65–175 × 2.5–4.5 μm, with a heavy wall. Penicilli were typically biverticillate (Figure 1c). Metulae in whorls of two to six, which were usually rather appressed, sometimes slightly divergent when forced apart by larger whorls, about 11.0–15.3(−16.0) × (2.0−)2.5–3.5(−4.0) μm across the top, individually more or less cylindrical. Phialides were acerose, 11.0–16.0 × 1.8–3.0(−3.5) μm, conidiogenous aperture 0.5–1.5 μm wide. Conidia borne in chains, were globose to subglobose, smooth-walled or slightly roughened, 2.0–4.0 × 2.0–3.7 μm.
The total length of the rDNA ITS (including 5.8S rDNA) of the strain FKI-5653 was 555 bp. In a BLAST search using blastn from the National Center for Biotechnology Information,17 the strain FKI-5653 was a 100% match to the nucleotide sequences of Penicillium pinophilum (GenBank accession number AY787845).
From the results of morphological characteristics and BLAST search, the strain FKI-5653 was identified as Penicillium pinophilum.
Taxonomy of strain FKI-5655
Colonies on PDA were >80 mm in radius after 7 days at 25 °C (Figure 2a), floccose, flat, with white (a) aerial mycelium, covered with dusty olive (1 lg) conidia, exudate lacking, soluble pigment not produced. Colonies on CMD were >80 mm in radius after 7 days at 25 °C, flat, the surface with ivory (2 db) floccose aerial mycelium, covered with dusty olive (1 lg) conidia, exudate lacking, soluble pigment not produced. Colonies on SNA were >80 mm in radius after 7 days at 25 °C, floccose, flat, with white (a) aerial mycelium, covered with light olive drab (1 li) conidia, exudate lacking, soluble pigment not produced. They were conidiated after 2 days in pustules more or less regularly, and pustules were distributed on the plate or forming in a broad band around the margin. Pustules were at first white, becoming green from the fourth day or later. The culture on PDA at 5 and 37 °C showed no growth.
Conidiophores were highly uniformly branched, branches frequently paired or in threes, arising at or near 90°, with respect to the main axis, longer and more profusely branched with distance from the tip (Figure 2b). Phialides were arising singly from main axis and branches, or held at or near 90° in whorls, tending to be conspicuously swollen below the sharply constricted tip, ampulliform, (6.0−)7.3–9.7(−13.0) μm long, (1.5−)2.0–3.0 μm at the widest point, (0.8−)1.0–1.5(−1.7) μm at the base. Conidia were globose to subglobose, 3.0–4.3(−4.8) × 2.8–4.3 μm. Chlamydospores were scattered, not abundant, terminal and intercalary in hyphae, globose or subglobose.
The total length of the rDNA ITS (including 5.8S rDNA) of the strain FKI-5655 was 570 bp. In TrichOKEY of International Subcommission on Trichoderma and Hypocrea taxonomy (ISTH),18 the query sequence of the strain FKI-5655 belongs to Trichoderma harzianum, and in TrichoBLAST of ISTH,18 the strain FKI-5655 was a 99.8% match to the nucleotide sequences of Trichoderma harzianum DAOM 231412.
From the results of the morphological characteristics, TrichOKEY and TrichoBLAST search, strain FKI-5655 was identified as Trichoderma harzianum.
Co-culture
The production of red pigments was observed in contact face on PDA plate only by co-culture of strains Penicillium pinophilum FKI-5653 and Trichoderma harzianum FKI-5655, but not by pure culture of each of them (Figure 3). As for liquid culture, co-culture in glucose–peptone broth showed more dark red than pure culture. This phenomenon illustrated an increasing diversity of metabolites in co-culture, and these were subsequently analyzed.
Isolation
The 6-day-old co-culture broth of the first batch (8 l) was centrifuged. Acetone (3 l) was added to the mycelium, centrifuged (3000 r.p.m.) and removed acetone in vacuo. The aqueous solution was extracted with ethyl acetate four times, followed by concentration of the organic layer in vacuo. From the extracts (2.3 g) of ethyl acetate, 1.8 g was applied on a silica gel column (55φ × 60 mm, Merck Co., Whitehouse Station, NJ, USA). After washing with CHCl3-MeOH (10:1, 360 ml), the CHCl3-MeOH (1:1, 360 ml) eluate was concentrated in vacuo to yield a crude material (303 mg). The crude material (280 mg) was re-chromatographed by silica gel column (10φ × 400 mm). After washing with CHCl3-MeOH (10:1, 100 ml), the CHCl3-MeOH (1:1, 100 ml) eluate was concentrated in vacuo to yield a crude material (163 mg). It was applied on an ODS column (18φ × 35 mm, Senshu Scientific Co., Tokyo, Japan) equilibrated with 50% MeOH aq. After washing with 50% MeOH aq (60 ml), secopenicillide C (1, 8 mg) was eluted with 55% MeOH aq. Then, followed by washing with 60% MeOH aq (60 ml); stromemycin (5, 2.7 mg) was eluted with 70% MeOH aq.
The 6-day-old co-culture broth of the second batch (5 l) was centrifuged. EtOH (5 l) was added to the culture broth, centrifuged (3000 r.p.m.) and removed EtOH in vacuo. The aqueous solution was extracted with ethyl acetate four times, followed by concentration in vacuo. From the extracts (3.0 g) of ethyl acetate, 2.5 g was applied on a silca gel column (30φ × 430 mm, Merck Co.) and eluted with CHCl3-MeOH (100:0, 100:1, 50:1, 10:1, 1:1, 0:100, each 900 ml). CHCl3-MeOH (400 mg; 50:1) eluate (450 mg) was applied on the second silica gel column (12φ × 400 mm) and eluted with CHCl3-MeOH (100:0, 100:1, 50:1, 10:1, 1:1, 0:100, each 150 ml). The CHCl3-MeOH (100:1 and 50:1) eluate (250 mg) was applied on an ODS column (18φ × 35 mm, Senshu Scientific Co.) and eluted with MeOH-H2O (50:50, 60:40, 70:30, 80:20, 90:10, 100:0, each 180 ml). The 50% MeOH aq fraction (200 mg) was re-chromatographed by an ODS column (18φ × 35 mm) eluting with MeOH-H2O (40:60, 45:55, 50:50, 55:45, 60:40, 65:35, 70:30, 80:20, 100:0, each 180 ml). MC-141 (4, 12.5 mg) was eluted with 45% MeOH aq. Other fractions were combined and re-chromatographed by an ODS column (18φ × 35 mm) eluting with MeOH-H2O (40:60, 45:55, 50:50, 55:45, 60:40, 65:35, 70:30, 80:20, 100:0 each 50 ml). Pestalasin A (2, 3.2 mg) was eluted with 40% MeOH aq. The CHCl3-MeOH (100:1) eluate (490 mg) of the first silica-gel chromatography was purified by HPLC on a Pegasil ODS column (20φ × 250 mm, Senshu Scientific Co.) with 65% MeOH aq at 7.0 ml min−1 detected at UV 210 nm. The peak at retention times of 48 min was collected and concentrated in vacuo to dryness to afford penicillide (3, 66 mg).
Structure elucidation
The molecular formulae of 2, 3, 4 and 5 were elucidated by HR-ESI-MS to be C14H16O5, C21H24O6, C21H22O6 and C38H48O12, respectively. Through comparison with reported data, 2, 3, 4 and 5 were identified as pestalasin A,19 penicillide,20, 21 MC-14122 and stromemycin,23 respectively.
Compound 1 was obtained as a light redish amorphous solid (UV (MeOH) λmax (log ɛ): 209 (5.51), 255 (5.04) and 300 (4.80)). The IR spectrum showed characteristic absorptions at 3423, 2956 and 1602 cm−1, suggesting the presence of hydroxy groups and aromatic rings. The molecular formula of 1 was elucidated by HR-ESI-MS to be C20H22O6 (found, 381.1297 [M+Na]+; calcd., 381.1314 for C20H22O6Na), requiring 10 degrees of unsaturation. The 1H and 13C NMR spectral data of 1 in CD3OD are listed in Table 1. The 13C NMR and HSQC spectra indicated 20 carbons, which were classified into one carboxyl carbon at δc 175.6, nine sp2 quaternary carbon at δc 159.9, 157.7, 150.7, 140.3, 136.7, 136.3, 132.5, 123.8 and 110.5, five sp2 methine carbon at δc 132.3, 123.9, 120.7, 117.9 and 106.5, two sp3 methylene carbons at δc 60.5 and 30.7, and three methyl carbons at δc 25.9, 21.4 and 17.8. As shown by the bold lines for 1 in Figure 4, proton spin networks from H2-1′ (δH 3.21) to H-2′ (δH 5.26) and from H-1 (δH 6.03) to H-2 (δH 6.89) were shown by the 1H-1H COSY. The correlations in HMBC spectra showed 1 has an isoprenyl group, a 1,2,3,4-tetrasubstituted benzene ring and a 1,2,3,5-tetrasubstituted benzene ring, which suggested 1 is an analog of penicillide (3) as shown in Figure 4. On the basis of the HMBC experiments, the key correlations from H2-1′ to C-4 (δc 159.9) and from H-2 to C-1′ (δc 30.7) indicated the isoprenyl group is linked to C-3. The HMBC correlations from H2-15 (δH 4.50) to C-8 (δc 140.3) and C-9 (δc 136.3) indicated the oxygenated methylene group is linked to C-9. Thus, the structure of 1 was elucidated as shown in Figure 5. Though 1 was listed in CAS Registry file (Registry number 914071-55-9) and ChemBank (ID 3553199), there were no published data. As seco-type compounds of penicillides were reported as secopenicillides A and B, 1 was desiganted secopenicillide C.
Comparison of metabolites productivity between pure culture and co-culture
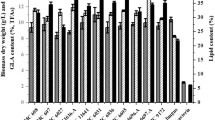
All isolated compounds were detected in the pure culture broth of Penicillium pinophilum FKI-5653 (Figure 6). The productivity of isolated compounds between co-culture and pure culture is shown in Figures 6 and 7. The productivity of penicillide, secopenicillide C, MC-141 and stromemycin in co-culture was 2.0-, 3.7-, 5.8- and 4.4-fold higher than that in pure culture of Penicillium pinophilum, respectively. On the contrary, the productivity of pestalasin A was 2.3-fold less than that of pure culture.
Discussion
We have isolated a new compound, designated secopenicillide C and four known compounds, penicillide, MC-141, pestalasin A and stromemycin, from the co-culture broth of Penicillium pinophilum FKI-5653 and Trichoderma harzianum FKI-5655. The production of these compounds except pestalasin A was enhanced in co-culture to be 2–6 times higher than in that pure culture of Penicillium pinophilum FKI-5653, which was a producer of these compounds. On the contrary, metabolites produced by Trichoderma harzianum FKI-5655 were reduced by co-culture, although it produced only small amount of metabolites in pure culture (Figure 6). We think this nature of Trichoderma harzianum FKI-5655 might be a great advantage to improve metabolites productivity of another fungi in co-culture. We are studying the factor of metabolites productivity enhancement now. Co-culture studies with this strain, together with other fungi, are in progress to test our hypothesis.
References
Chin, Y. W., Balunas, M. J., Chai, H. B. & Kinghorn, A. D. Drug discovery from natural sources. AAPS J. 8, E239–E253 (2006).
Chiang, Y. M. et al. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem. Biol. 15, 527–532 (2008).
Onaka, H., Mori, Y., Igarashi, Y. & Hurumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 77, 400–406 (2011).
Kurosawa, K. et al. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J. Am. Chem. Soc. 130, 1126–1127 (2008).
Schroeckh, V. et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl Acad. Sci. USA 106, 14558–14563 (2009).
Zuck, K. M., Shipley, S. & Newman, D. J. Induced production of N-formyl alkaloids from Aspergillus fumigatus by co-culture with Streptomyces peucetius. J. Nat. Prod. 74, 1653–7 (2011)(in press).
Cueto, M. et al. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 64, 1444–1446 (2001).
Park, H. B., Kwon, H. C., Lee, C. H. & Yang, H. O. Glionitrin A, an antibiotic-antitumor metabolite derived from competitive interaction between abandoned mine microbes. J. Nat. Prod. 72, 248–252 (2009).
Gibson, J., Sood, A. & Hogan, D. A. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 75, 504–513 (2009).
Degenkolb, T., Heinze, S., Schlegel, B., Strobel, G. & Grafe, U. Formation of new lipoaminopeptides, acremostatins A, B, and C, by co-cultivation of Acremonium sp. Tbp-5 and Mycogone rosea DSM 12973. Biosci. Biotechnol. Biochem. 66, 883–886 (2002).
Sonnenbichler, P., Dietrich, J. & Peipp, H. Secondary fungal metabolites and their biological activities, V. Investigations concerning the induction of biosynthesis of toxic secondary metabolites in basidiomycetes. Biol. Chem. 375, 71–79 (1994).
Zhu, F. & Lin, F. Marinamide, a novel alkaloid and its methyl ester produced by the application of mexed fermentation technique to two mangrove endophytic fungi from the South China Sea. Chin. Sci. Bull. 51, 1426–1430 (2006).
Zhu, F., Lin, Y., Wang, X. & Huang, L. Secondary metabolites of two marine-derived mangrove endophytic fungi (strain nos. 1924 and 3893) by mixed fermentation. Chem. Ind. Forest Prod. 27, 8–10 (2007).
Pitt, J. I. The Genus Penicillium, and its Teleomorphic States Eupenicillium and Talaromyces. pp. 1–634 (Academic Press: London, 1979).
Jacobson, E., Granville, W. C. & Foss, C. E . Color Harmony Manual 4th edn (Container of America: Chicago, 1958).
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, In: PCR Protocols: a Guide to Methods and Applications (eds Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J.) (Academic Press: New York, 315–332 1990).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Druzhinina, I. et al. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fung. Gen. Biol. 42, 813–828 (2005).
Xu, J. et al. Cytosporones, Coumarins, and an alkaloid from the endophytic fungus Pestalotiopsis sp. Isolated from the Chinese mangrove plant Rhizophora mucronata. Bioorg. Med. Chem. 17, 7362–7367 (2009).
Xia, X. K. et al. 1H and 13C NMR assignments for 6-demethylvermistatin and two penicillide derivatives from the mangrove fungus Guignardia sp. (No. 4382) from the South China Sea. Magn. Reson. Chem. 46, 693–696 (2008).
Komai, S. et al. New penicillide derivatives isolated from Penicillium simplicissimum. J. Nat. Med. 60, 185–190 (2006).
Suzuki, K., Nozawa, K., Udagawa, S., Nakajima, S. & Kawai, K. Penicillide and dehydroisopenicillide from Talaromyces derxii. Phytochemistry 30, 2096–2098 (1991).
Bringmann, G., Lang, G., Steffens, S., Gunther, E. & Schaumann, K. Evariquinone, isoemericellin, and stromemycin from a sponge derived strain of the fungus Emericella variecolor. Phytochemistry 63, 437–443 (2003).
Acknowledgements
This study was supported, in part, by funds from Quality Assurance Framework of Higher Education from the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT). We are grateful to Ms Akiko Nakagawa, Dr Kenichiro Nagai and Ms Noriko Sato, School of Pharmacy, Kitasato University, for measurements of mass and NMR spectra.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Nonaka, K., Abe, T., Iwatsuki, M. et al. Enhancement of metabolites productivity of Penicillium pinophilum FKI-5653, by co-culture with Trichoderma harzianum FKI-5655. J Antibiot 64, 769–774 (2011). https://doi.org/10.1038/ja.2011.91
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.91
Keywords
This article is cited by
-
Solid-state co-culture fermentation of simulated food waste with filamentous fungi for production of bio-pigments
Applied Microbiology and Biotechnology (2022)
-
Co-Culture of Plant Beneficial Microbes as Source of Bioactive Metabolites
Scientific Reports (2017)
-
Antibiotics in microbial coculture
The Journal of Antibiotics (2017)
-
Coculnol, a new penicillic acid produced by a coculture of Fusarium solani FKI-6853 and Talaromyces sp. FKA-65
The Journal of Antibiotics (2015)
-
Induced production of BE-31405 by co-culturing of Talaromyces siamensis FKA-61 with a variety of fungal strains
The Journal of Antibiotics (2015)