Abstract
A simple and rapid screen was performed for microorganisms producing cyclic lipopeptide acylase, an enzyme that deacylates the acyl side chain of FR901379 to yield a cyclic peptide, FR179642, which is an important intermediate in producing micafungin. Among ∼3800 newly isolated strains from soil samples, 5 microorganisms expressing high acylase activity were selected and classified, 3 as Streptomyces spp. and 2 as filamentous fungi. This is the first time that these strains have been identified as cyclic lipopeptide acylase producers. Both fungi and streptomycetes showed differing optimum pH and temperature profiles for acylase activity.
Similar content being viewed by others
Introduction
FR901379,1 a product of the fungus Coleophoma empetri F-11899, is an echinocandin-like compound that inhibits the synthesis of 1,3-β-D-glucan in the fungal cell wall. This compound has good water solubility due to sulfate ester moiety on the homophenylalanine group, whereas other echinocandin-like compounds, such as echinocandin B, aculeacin, mulundocandin and pneumocandin are barely water soluble. This insolubility is one of the reasons why they have not been developed for clinical use. In contrast, FR901379 shows significant hemolytic activity as a result of its long acyl chain and is therefore also inappropriate for clinical use. Therefore, we attempted to produce improved derivatives by substituting the acyl chain; to achieve this, production of a key intermediate, FR179642 or deacylated FR901379, was critical. Acyl peptides are generally unstable to chemical deacylation and therefore enzymatic deacylation is widely applied. Therefore, we used bioconversion to obtain FR179642 (Figure 1). In the past, deacylated products were produced by enzymatic deacylation using penicillin G acylase,2 cephalosporin C acylase,3 echinocandin B acylase4 or aculeacin A acylase.5 At first, we used acylase produced by Actinoplanes utahensis that was well known for removing acyl side chains from echinocandins. However, we had to change the procedure to use a more specific acylase with higher potency toward FR901379 because a large quantity of the deacylated FR901379 was required to synthesize the derivatives for optimizing the side chain. Therefore, we undertook a screening process to identify a novel lipopeptide acylase to remove the acyl side chain (palmitoyl) from FR901379 and, as a result, discovered several strains that showed this acylase activity.
In this study, we describe the screening process for FR901379 acylase-producing strains and their subsequent taxonomic characterization, as well as some comparisons of enzymatic activity with the known lipopeptide acylase-producing microorganism.
Materials and methods
Reagents and microorganisms
FR901379 and echinocandin B were prepared from the fermentation broth of C. empetri F-118991 and Aspergillus nidulans var. echinulatus A-32204, NRRL 3860,6 respectively. Other reagents were purchased from Wako Pure Chemical Industries (Osaka, Japan).
Screening for FR901379 acylase-producing microorganisms
Microorganisms
Approximately 3300 actinomycetes and 500 fungi isolated from soil samples collected from various parts of Japan and stored at Fermentation Research Laboratories, Astellas Pharma (Aichi, Japan) were examined.
Assay method
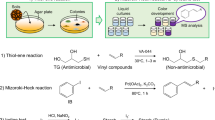
As the first screening step, microorganisms were grown on agar supplemented with 1 mg ml−1 of FR901379. Once colonies reached a diameter of 5–10 mm, the plate was overlaid with agar medium containing 1 × 103 per ml cells of Candida albicans Yu-1200 and incubated at 30 °C for 16 h. The candidate microorganism producing FR901379 acylase was identified as the colony allowing growth of C. albicans (Figure 2). This method is based on the observation that C. albicans is sensitive to FR901379, but is insensitive to its deacylated compound FR179642. The second step consisted of enzymatic hydrolysis of FR901379 by the cultured broth of the candidate microorganism, and determination of the production of FR179642 by HPLC.
For the second screening step, actinomycetes were inoculated into the medium (glucose 4%, yeast extract 1%, KH2PO4 0.1%, pH adjusted to 6.5 before sterilization), whereas fungi were inoculated into the same medium supplemented with 1 mg ml−1 of sterilized FR901379, as some of the tested fungal strains showed deacylation activity only when cultured with a medium supplemented with FR901379. Inoculated media were cultured at 25 °C for 2–5 days until microbes were well grown in the liquid culture.
The reaction mixture for enzymatic hydrolysis contained 1 mg ml−1 FR901379, 50 mM phosphate buffer (pH 7.0) and cultured broth in a final volume of 1 ml. The mixture was incubated at 30 °C for 1 day and centrifuged at 2000 r.p.m. for 20 min. The supernatant was analyzed by HPLC (Shimadzu Corporation, Kyoto, Japan) to detect deacylated FR901379 (FR179642). The HPLC system comprised a LiChrospher 100RP-18 column (250 mm × 4 mm i.d.) (Merck KGaA, Darmstadt, Germany) and a mobile phase of 5% acetonitrile/0.5% ammonium dihydrogenphosphate, with a UV detector at 210 nm and a flow rate of 1 ml per min. The retention time of FR179642 was ∼6.3 min.
Taxonomic studies
Cultural characteristics were observed on several media incubated at 30 °C for 14 days and at 25 °C for 14 days for actinomycetes and fungi, respectively. Color names used in this study were taken from the Methuen Handbook of Colour.7
Results
Screening for FR901379 acylase-producing microorganisms
A total of 145 strains were selected in the first screening step. Among these, 22 actinomycetes and 2 fungi were confirmed to produce the deacylated compound FR174692 by HPLC. In all, three strains of actinomycetes, that is, no. 4811, no. 6907 and no. 8703, which showed relatively higher activity than others, and two fungi, no. 30 084 and no. 30 085, were chosen for further characterization.
Characterization of FR901379 acylase-producing strains
Strain no. 4811, no. 6907 and no. 8703 were isolated from a soil sample collected in Fukushima Prefecture. The cultural characteristics on yeast extract–malt extract agar, oatmeal agar, inorganic salts–starch agar, glycerin–asparagine agar, peptone–yeast extract–iron agar and tyrosine agar, as well as the light and scanning electron microscopy observations of the respective growths are summarized in Table 1.
For both no. 4811 and no. 8703, the color of the aerial mycelium was yellowish gray to greenish gray, the reverse side color of growth was yellowish brown to brown, the soluble pigment was light brown and neither intracellular pigments nor soluble pigments were pH sensitive. No melanoid pigments were produced in strain no. 4811, whereas melanoid pigments were produced in no. 8703 in the tryptone–yeast extract broth, peptone–yeast extract–iron agar and tyrosine agar. The color of the aerial mycelium of strain no. 6907 was yellowish gray to bluish gray, the reverse side color of growth was light brown to brown and the intracellular pigments were not pH sensitive. Melanoid pigments were produced in tryptone–yeast extract broth, peptone–yeast extract–iron agar and tyrosine agar.
The physiological characteristics are summarized in Table 2. Strain no. 4811 and no. 8703 did not use inositol, sucrose and raffinose but peptonized milk; the temperature range for growth was 4.0–35.0 °C. In contrast, strain no. 6907 used all carbon sources tested but did not peptonize milk; the temperature range for growth was 9.0–40.0 °C.
The vegetative mycelium of strain no. 4811, no. 6907 and no. 8703 developed well with irregular, but not fragmented, branching. The aerial mycelium extending from the vegetative mycelium branched monopodially to form elongated spore chains. The spore chain morphology of the aerial mycelium of strain no. 4811 and no. 8703 was straight to flexuous, thus belonging to the RF type according to the classification described by Pridham et al.8 The spore chain morphology of the aerial mycelium of no. 6907 was straight to flexuous or incomplete loops, thus belonging to the RF or RA types. Each spore chain consisted of ⩾20 spores. The spores were smooth-surfaced (glabrous) and cylindrical. The spore size was 0.5–0.7 × 0.7–1.1 μm2 for no. 4811, 0.5–0.7 × 0.7–1.3 μm2 for no. 6907 and 0.5–0.3 × 0.6–1.1 μm2 for no. 8703. No sclerotium, sporangium or zoospore were observed.
With regard to the amino-acid composition of the cell wall, the whole-cell lysate was analyzed by methods described by Becker et al.9 and Yamaguchi10 with results demonstrating LL-diaminopimelic acid in all strains. Therefore, the cell wall of these three strains was considered to belong to type 1.
On the basis of the above-mentioned morphological observations and chemical analysis, strain no. 4811, no. 6907 and no. 8703 were considered to belong to the genus Streptomyces according to the taxonomic classification described by Pridham et al.8 Accordingly, the characteristics of these strains were compared with those of other Streptomyces species as described in the literature, namely by Shirling et al.,11, 12, 13 Skerman et al.14 and Moore et al.15 These comparisons indicated that the described characteristics of Streptomyces anulatus were substantially identical to those of strain no. 4811 and no. 8703, and therefore they were identified and named S. anulatus no. 4811 and S. anulatus no. 8703, respectively. Comparisons with no. 6907 were unable to identify a specific species for the strain, and it was therefore named Streptomyces sp. no. 6907.
S. anulatus no. 4811, S. anulatus no. 8703 and Streptomyces sp. no. 6907 were originally deposited with the National Institute of Bioscience and Human Technology (NIBH) under the accession numbers FERM P-15377, FERM P-15507 and FERM P-15506, respectively, and subsequently converted to the accession numbers FERM BP-5808, FERM BP-5810 and FERM BP-5809, respectively, according to the Budapest Treaty.
Fungal strain no. 30 084 and no. 30 085 were isolated from a sample collected in Jouhoku-machi, Higashi Ibaraki-gun (Ibaraki, Japan). These strains showed restricted growth on various media and formed colonies varying in color according to the different type of culture media used: for example, greenish gray, brownish orange, yellowish white for no. 30 084, greenish gray, yellowish gray, brown, etc. and brown soluble pigments diffusing into the medium for no. 30 085. On several media, strain no. 30 084 formed anamorphs showing a conidial structure consisting of a dendritic conidiophore rising up from the surface of the medium with arthroconidia at its branches. Although no sporogenic organ was observed normally in no. 30 085, this strain formed a minimal quantity of asexual spores after prolonged incubation on corn meal agar.
The cultural characteristics of these fungi on various agar media are summarized in Table 3. Colonies of no. 30 084 on malt extract agar grew regressively and spread to attain diameters of 1.5–2.0 cm after 2 weeks of incubation at 25 °C. The colony was circular and was either raised as a whole or elevated peripherally and depressed in the center. The strain formed anamorphs in abundance, presenting a powdery surface. The colony was greenish gray with a yellowish-gray peripheral zone and the reverse side was light yellow with a yellowish-white peripheral zone. On potato dextrose agar, the colony also showed restricted growth and spread to attain diameters of 1.5–2.0 cm under the same cultural conditions as described above. The surface of the colony was centrally elevated or convex, wrinkled and somewhat felt-like, forming a small amount of anamorphs. The colony was brownish orange to light brown with an orange-white peripheral zone, and the reverse side was brown with an orange-white peripheral zone.
The morphological characteristics of strain no. 30 084 were recorded by observing its growth in Miura's medium.16 The conidiophore of strain no. 30 084 stood erect from the surface of the medium and consisted of a tan-colored linear trunk with colorless intricate branches. This dendritic structure made the conidiophore clearly distinguishable from the vegetative hypha. The conidiophore was 90–220 (or 240) μm in height, with its trunk being 2–3 (or 3.5) μm wide. The branches were bifurcated or trifurcated in succession and spread to occupy the space 30–50 μm in height and 40–60 μm in width over the top of the conidiophore. Each branch was fragmented along multiple septae, forming conidia in either form of a rod or an ellipsoid truncated at one or both ends. The conidium was colorless, smooth-surfaced, unicellular and varied in size from 2–4 × 1.5–2 μm2. Individual conidia were linked through vestiges of cell walls of their branches. Vegetative hyphae were smooth-surfaced, septated, colorless and branched. The hyphal cell was cylindrical and 1.0–2.5 μm in width. No clamydospores were observed. Strain no. 30 084 could grow at 3–32 °C, and the optimum temperature for growth was 24–28 °C. The above-mentioned characteristics of strain no. 30 084 were observed after incubation at 25 °C and compared with descriptions in several books17, 18, 19 on the taxonomy of fungi. As a result, these characteristics were found to agree with descriptions of the genus Oidiodendron, and the strain was therefore identified as belonging to this genus and named Oidiodendron sp. no. 30 084. Although this strain was originally deposited with NIBH and assigned the accession number FERM P-15550, it was subsequently converted to a deposit according to the Budapest Treaty and assigned the accession number FERM BF-5943.
The colony of strain no. 30 085 on potato dextrose agar showed restricted growth and spread to attain diameters of 2.0–3.0 cm after 2 weeks of incubation at 25 °C. The colony was circular, flat-to-elevated, slightly wrinkled and felt-like. An orange-colored exudate was observed on the surface of the colony, with the colony itself being greenish gray to pale gray with an orange white to orange-gray peripheral zone. The reverse side was grayish brown to dark brown with a grayish-orange peripheral zone and a diffusion of brown soluble pigments into the medium. The colony on Czapek's solution agar showed markedly restricted growth on Czapek's solution agar and spread circularly to attain diameters of 1.0–2.0 cm after 2 weeks of incubation at 25 °C. The surface of the colony was flat and thin, with no aerial hyphae. The surface color was white or yellowish gray with the reverse side being the same. The conidiophore of strain no. 30 085 was not clearly distinguishable from the vegetative and aerial hyphae, with 2–5 conidiogenous cells occurring in whirls, or at times singularly, on the side of the vegetative hyphae. Conidiogenous cells were colorless, smooth-surfaced, filamentous to elongated flask-shaped (lecythiform), measuring 18–37 (at times 45) × 1.5–2.0 μm2 and formed a single conidium to several conidia in a continuous series at the tip. The mode of conidiogenesis appeared to be phialidic but no definite collarette was observed. The conidia were colorless, smooth-surfaced, prolate (ellipsoidal) to bacilliform (rod-shaped), unicellular and 3–5.5 × 1.5–2.5 μm2 in size. Vegetative hyphae were smooth-surfaced, septated, colorless and branched, and although they were usually linear, at times they could also be curved and remarkably crimped. The hyphal cell was cylindrical to filiform and 1.0 × 5.0 μm2 in width, containing a large number of intracellular vacuoles. These vacuoles were released extracellularly with cell aging to yield a highly viscous exudate. Although clamydospores, sclerotia and catenulate forms were not observed, in old cultures the pleurogenesis of a large number of globose cells was observed. Strain no. 30 085 was able to grow at 4–29 °C, and the optimum temperature for growth was 22–26 °C. on potato dextrose agar. The above-mentioned characteristics of strain no. 30 085 were observed after 14 days of incubation at 25 °C and compared with the descriptions in several books20, 21, 22 on the taxonomy of fungi. As a result, strain no. 30 085 was found to resemble Verticillium. Although fungi of the genus Verticillium typically produce a large number of conidia as a mucoid mass at the tip of the phialide, we considered the inability of strain no. 30 085 to produce numerous conidia to be due to its poor sporogenous ability, and therefore identified this strain as a species of the genus Verticillium and named it Verticillium sp. no. 30 085. Although this strain was originally deposited with NIBH, assigned the accession number FERM F-15551 and designated as fungus no. 30 085, it was subsequently renamed Verticillium sp. no. 30 085 and converted to a deposit according to the Budapest Treaty with the accession number FERM BP-5944.
Optimum pH and temperature for acylase activity and substrate specificity
The optimum pH and temperature for acylase activity against FR901379 were determined using the culture broth of each selected strain. Profiles of optimum pH and temperature for acylase activity that differed greatly between streptomycetes and fungi are shown in Figure 3. The acylase activity of fungi occurred at a lower pH and temperature than that of Streptomycetes. Streptomyces no. 6907 showed the most potent acylase activity and was consequently compared with A. utahensis NRRL12052 against FR901379 and echinocandin B. In this protocol, 20 ml of cultured broth of Streptomyces no. 6907 and A. utahensis NRRL12052 was prepared as described in the literature23 and 1 ml of each substrate solution (200 mg m1−1) together with 2.9 g of disodium hydrogen phosphate and 60 ml of water were added. The reaction was carried out at 60 °C for 24 h. After completion, the reaction mixture was filtered through a membrane filter (0.45 mm) to remove high molecular proteins. and each deacylated product was quantified by HPLC. As shown in Table 4, the acylase activity of Streptomyces no. 6907 was more potent and specific to FR901379 than that of A. utahensis NRRL12052.
Discussion
Deacylation of microbial products is essential for the production of a number of important medicines. We used microbial deacylation to semi-synthesize a novel lipopeptide antifungal drug for deep-seated mycoses, through enzymatic deacylation of FR901379, followed by chemical reacylation with an optimized acyl group.24 We required our original acylase to show potent activity against FR901379 for obtaining a large quantity of deacylated compound during the optimization study and for establishing industrial production. As A. utahensis produces an enzyme to deacylate several lipopeptide compounds, we searched for novel acylase-producing microorganisms among wild actinomycetes and fungal strains freshly isolated from soil samples. Consequently, three Streptomyces and two filamentous fungal strains were isolated. Taxonomic studies have shown that two of the three Streptomycetes strains were species of S. anulatus and that each of the two fungal strains belonged to the genera Verticillium and Oidiodendron. Although it has been previously reported that A. utahensis,4, 5 S. mobaraensis25 and Streptomyces sp.26 produce acylases, our study is the first to find that S. anulatus also produces an acylase. Elucidation of the protein structure for acylases produced by these newly identified strains will be reported elsewhere, together with comparison of the amino-acid sequences with that of known acylases.
All acylases so far derived from actinomycetes are reported to belong to the family of penicillin acylases that have a heterodimer protein structure. Our preliminary data indicated that Streptomyces strains obtained in this study produce acylases with the same properties as those of penicillin acylase. In contrast, data regarding optimum pH and temperature for acylase activity suggested that fungal acylases have different properties from acylases produced by Streptomyces. Further, although penicillin acylases are constitutively produced, the addition of FR901379 to culture media was necessary to induce acylase activity in fungi. We are presently undertaking a detailed analysis on the structure and enzymology of these new acylases.
Conflict of interest
The authors declare no conflict of interest.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Iwamoto, T. et al. WF11899A, B and C, novel antifungal lipopeptides. I. Taxonomy, fermentation, isolation and physico-chemical properties. J. Antibiot. 47, 1084–1091 (1994).
Kleinshmidt, W. J., Wright, W. E., Kavanagh, F. W. & Stark, W. M. (Eli Lilly & Co.). Penicillin Deacylation Via Actinoplaneceae fermentation, U.S. 3,150,059, September 22 (1964).
Ishii, Y. et al. High-level production, chemical modification and site-directed mutagenesis of a cephalosporin C acylase from Pseudomonas strain N176. Eur. J. Biochem. 230, 773–778 (1995).
Boeck, L. D., Fukuda, D. S., Abbott, B. J. & Debono, M. Deacylation of echinocandin B by Actinoplanes utahensis. J. Antibiot. 42, 382–388 (1989).
Debono, M. et al. A21978C, a complex of new acidic peptide antibiotics: isolation, chemistry, and mass spectral structure elucidation. J. Antibiot. 40, 761–777 (1989).
Benz, F., Knusel, F., Nuesch, J., Treichler, H. & Voser, W. Echinocandin B, a new polypeptide antibiotic from Aspergillus nidulans var. echinulatus. Helv. Chim. Acta. 57, 2459–2477 (1974).
Wanscher, J. H. & Kornerup, A. Methuen Handbook of Colour 3rd edn, (Eyre Methuen, London, 1978).
Pridham, T. G., Hesseltine, C. W. & Benedict, R. G. A guide for the classification of streptomycetes according to selected groups; placement of strains in morphological sections. Appl. Microbiol. 6, 52–79 (1958).
Becker, B., Lechevalier, M. P., Gordon, R. E. & Lechevalier, H. A. Rapid differentiation between Nocardia and Streptomyces by paper chromatography of whole-cell hydrolysates. Appl. Microbiol. 12, 421–423 (1964).
Yamaguchi, T. Comparison of the cell-wall composition of morphologically distinct actinomycetes. J. Bacteriol. 89, 444–453 (1965).
Shirling, E. B. & Gottlieb, D. Cooperative description of type culture of Streptomyces. 2. Species descriptions from first study. Intern. J. Syst. Bacteriol. 18, 69–189 (1968).
Shirling, E. B. & Gottlieb, D. Cooperative description of type culture of Streptomyces. 3. Additional species descriptions from first and second studies. Intern. J. Syst. Bacteriol. 18, 279–392 (1968).
Shirling, E. B. & Gottlieb, D. Cooperative description of type culture of Streptomyces. 4. Species descriptions from second, third and forth studies. Intern. J. Syst. Bacteriol. 19, 391–512 (1969).
Skerman, V. B., McGowan, V. & Sneath, P. H. A. Approved List of Bacterial Names (Amended Edition, American Society for Microbiology, Washington D.C., 1989).
Moore, W. E., Cato, E. P. & Moore, L. V. H. Index of bacterial and yeast nomenclatural changes (American Society for Microbiology, Washington D. C., 1992).
Miura, K. & Kudo, M. An agar-medium for aquatic hyphomycetes. Trans. Mycol. Soc. Japan. 11, 116–118 (1970).
Barron, G. R. The Genera of Hyphomycetes from Soil 239–241 (Williams & Wilkins, Baltimore, 1968).
Arx, J. A. The Genera Of Fungi. Sporulating in Pure Culture 180–184 (J. Cramer, Vaduz, 1974).
Domson, K. H., Gams, W. & Anderson, T. H. Compendium of Soil Fungi 517–524 (Academic Press, London, 1980).
Barron, G. R. The Genera of Hyphomycetes from Soil 364 (Williams & Wilkins, Baltimore, 1968).
Arx, J. A. The Genera of Fungi, Sporulating in Pure Culture 315 (J. Cramer, Vaduz, 1974).
Domsch, K. H., Gams, W. & Anderson, T. H. Compendium of Soil Fungi 859 (Academic Press, London, 1980).
Boeck, L. D., Fukuda, D. S., Abbott, B. J. & Debono, M. Deacylation of A21978C, an acidic lipopeptide antibiotic complex, by Actinoplanes utahensis. J. Antibiot. 41, 1085–1092 (1988).
Barrett, D. From natural products to clinically useful antifungals. Biochim. Biophys. Acta. 1587, 224–233 (2002).
Zhang, D., Koreishi, M., Imanaka, H., Imamura, K. & Nakanishi, K. Cloning and characterization of penicillin V acylase from Streptomyces mobaraensis. J. Biotechnol. 128, 788–800 (2007).
Park, S. Y. et al. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71, 2632–2641 (2005).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ueda, S., Sakamoto, K., Oohata, N. et al. Screening and characterization of microorganisms with FR901379 acylase activity. J Antibiot 63, 65–70 (2010). https://doi.org/10.1038/ja.2009.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2009.119
Keywords
This article is cited by
-
Deacylation of Echinocandin B by Streptomyces species: a novel method for the production of Echinocandin B nucleus
3 Biotech (2019)
-
Microbial metabolism of quorum-sensing molecules acyl-homoserine lactones, γ-heptalactone and other lactones
Applied Microbiology and Biotechnology (2014)
-
Echinocandins: production and applications
Applied Microbiology and Biotechnology (2013)
-
Cloning and expression of the FR901379 acylase gene from Streptomyces sp. no. 6907
The Journal of Antibiotics (2011)