Abstract
Ion-translocating retinylidene rhodopsins are widely distributed among marine and freshwater microbes. The translocation is light-driven, contributing to the production of biochemical energy in diverse microbes. Until today, most microbial rhodopsins had been detected using bioinformatics based on homology to other rhodopsins. In the past decade, there has been increased interest in microbial rhodopsins in the field of optogenetics since microbial rhodopsins were found to be most useful in vertebrate neuronal systems. Here we report on a functional metagenomic assay for detecting microbial rhodopsins. Using an array of narrow pH electrodes and light-emitting diode illumination, we were able to screen a metagenomic fosmid library to detect diverse marine proteorhodopsins and an actinorhodopsin based solely on proton-pumping activity. Our assay therefore provides a rather simple phenotypic means to enrich our understanding of microbial rhodopsins without any prior knowledge of the genomic content of the environmental entities screened.
Similar content being viewed by others
Introduction
Microbial rhodopsins function as light-driven (retinal-based) membranal ion pumps, cation channels or light sensors (Heberle and Deupi, 2014). These rhodopsins are found in various microorganisms (Archaea, Bacteria, Eukarya) (Grote and O'Malley, 2011; Béjà et al., 2013; Ernst et al., 2014) and viruses (Yutin and Koonin, 2012; Philosof and Béjà, 2013), and are classified as type I rhodopsins (distinguished from type II animal visual rhodopsins (Spudich et al., 2000)). Microbial rhodopsin ion pumps function as light-driven proton pumps (for example, bacteriorhodopsins, proteorhodopsins and xanthorhodopsins (Oesterhelt and Stoeckenius, 1971; Béjà et al., 2000a; Balashov et al., 2005)), chloride pumps (archaeal halorhodopsins and bacterial chloride rhodopsins (Lanyi and Weber, 1980; Lanyi and Oesterhelt, 1982; Yoshizawa et al., 2014)) and bacterial sodium pumps (Inoue et al., 2013).
Current discoveries of novel microbial rhodopsin ion pumps are restricted to homology searches using known rhodopsins as genomic baits. Only one study used phenotypic characterization in order to detect proteorhodopsins in metagenomic libraries (Martínez et al., 2007). Martínez et al. screened some 12 000 marine environmental fosmid clones for the characteristic pinkish/orange color of E. coli colonies that express microbial rhodopsins. Three colonies were scored positive (based on their color), and they contained a proteorhodopsin gene as well as a gene cassette coding for a retinal biosynthetic pathway resembling the cassettes reported by Sabehi et al. (2005). We envisioned another phenotypic screen for microbial rhodopsins that would monitor ion-pumping activity directly upon illumination of different microbial rhodopsin transporters. Light-driven ion transport by rhodopsins can be measured directly with a pH electrode. Changes in pH indicate outward proton pumping when acidification of the medium is observed, but can also indicate inward chloride pumping or outward sodium pumping when alkalization of the medium is observed (see Inoue et al. (2014) for a schematic illustration of different light-driven pH changes behaviors of different proton, chloride or sodium rhodopsin transporters).
Here we report on an assay utilizing a simple device containing an array of narrow pH electrodes that could be illuminated from beneath using light-emitting diode (LED) illumination. Using this device, a subset of an environmental fosmid library from the Red Sea was screened (20 plates, 1920 clones) and 11 clones exhibited light-driven proton-pumping activity. All 11 clones contained a rhodopsin gene (diverse proteorhodopsins and a marine actinorhodopsin), with some fosmids also containing an adjacent gene cassette coding for a retinal biosynthetic pathway.
Results and discussion
A simple device containing eight narrow pH electrodes and LED illumination was designed, which would enable the direct screening of environmental fosmid libraries in a 96-well format (Figures 1a and b). To adjust our device, we used a positive clone (HF10_19P19) from the study of Martínez et al., 2007 that contained a proteorhodopsin gene and a retinal biosynthesis operon (Figure 1c). The use of narrow electrodes was essential for the simultaneous measurement of eight clones. An environmental fosmid library was constructed from Red Sea surface water (Supplementary methods), and copy control fosmids were used as cloning vectors to construct the library, as light-driven pH changes were observed only after the induction of the plasmid copy number from 1 to ~60 (data not shown).
Light-driven transport of protons in the Red Sea fosmid library. (a) LED setup. Upper panel, encasing of the LED light array; lower panel, close-up on the LED array. (b) Device setup. (c) pH changes in E. coli negative control (empty vector) and fosmid HF10_19P19 proteorhodopsin-expressing cells under dark (gray bars) and light (white bars). (d) Light-driven pH changes in the Red Sea fosmid library plate EIL80. Signals from positive wells are highlighted (wells are marked in bold). (e) Light-driven pH changes in positive clones EIL80B09 and EIL80E09. All assays were conducted in the presence of 10 μM all-trans retinal and under induced copy control fosmid conditions.
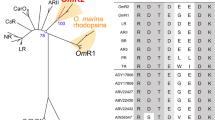
An example of the screen is shown for plate EIL80 (Figure 1d) under induced copy control conditions and with the addition of external all-trans retinal. As seen, light-driven proton-pumping activity was observed in two clones (EIL80B09 and EIL80E09), with clone EIL80E09 showing stronger proton-pumping activity (Figure 1e). The color of both clones’ pellets could not be distinguished clearly from the pale yellowish color of the negative control E. coli pellet (see the left panel of Figure 2a). Full sequencing of both fosmids revealed a proteorhodopsin gene on both, and a retinal biosynthetic pathway on clone EIL80B9 (Figure 2a).
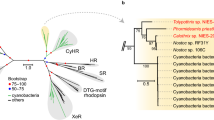
Red Sea microbial rhodopsins. (a) Schematic representation of the rhodopsin gene clusters identified in this work. Rhodopsin genes are marked in black, while genes in the retinal biosynthetic pathway are marked in green. Cell pellets (in the presence of retinal) are shown on the left. Predicted origin (shown on the right) is based on top blastX hits of different open reading frames on each clone (other than the rhodopsin and retinal biosynthetic pathway genes). (b) Unrooted maximum likelihood phylogenetic tree of microbial rhodopsin amino-acid sequences. Rhodopsins identified in this study are indicated with red circles. White circles represent bootstrap values >80%. Trees were constructed using the phylogeny.fr pipeline (Dereeper et al., 2008), which included PhyML v3.0 (Guindon et al., 2010) and the WAG substitution model for amino acids (Whelan and Goldman, 2001). One hundred bootstrap replicates were conducted for each analysis.
Screening overall 20 library plates (20 × 96=1920 clones), 11 clones showed light-dependent pH decrease when all-trans retinal was added to the growth medium (Supplementary Table). Light-dependent alkalization, indicative of potential chloride or sodium pumps, was not observed with any of the tested library plates. When retinal was omitted, only clone EIL102C09 retained activity. Clone EIL102C09 was also the only clone to show a colored (orange) cell pellet (left side of Figure 2a), possibly indicative of a strong expression of a rhodopsin gene in this clone. All 11 positive fosmids were fully sequenced and contained a microbial rhodopsin gene, with some containing a full or partial retinal biosynthetic pathway (Figure 2a). Four of the positive clones detected did not contain any retinal biosynthesis related genes. Ten of the identified rhodopsins were proteorhodopsins, while one was similar to rhodopsins from the recently reported uncultured low GC and ultra-small marine Actinobacteria (Ghai et al., 2013) (see phylogenetic tree in Figure 2b).
As observed in four of our clones, marine SAR86 bacteria (Dupont et al., 2012) and some freshwater Actinobacteria (Garcia et al., 2013; Keffer et al., 2015) lack a recognizable retinal biosynthesis pathway. These microbes seem to scavenge retinal or structurally related pigments from the environment in order to activate and utilize their rhodopsins (Béjà et al., 2001; Keffer et al., 2015). However, retinal concentrations in sea water and fresh water are unknown.
The host used in our screens is the gamma proteobacteria E. coli, and it was therefore not surprising to detect rhodopsins from predicted marine gamma proteobacterial groups (such as SAR86). In addition, our screen detected rhodopsins from various alpha groups (SAR11 and SAR116) and delta proteobacteria (SAR324), and even from a Gram-positive bacterial group (the low GC and ultra-small marine Actinobacteria). Our assay, therefore, seems elastic enough and is not restricted only to gamma proteobacteria-originating clones.
What does our screen miss?
Our designed screening system is based on the expression of microbial rhodopsins by their native promoters. Therefore, the following options are predicted to be not detected by our system: (i) promoters weakly recognized by E. coli will exhibit activity under our detection limits; (ii) promoters that are not recognized by E. coli will not be expressed; (iii) expressed but misassembled rhodopsins in E. coli membrane (for example, archaeal bacteriorhodopsins) will not exhibit measurable activity; (iv) every environmental DNA that is toxic to E. coli will be absent from our fosmid library (Béjà et al., 2000b; Sorek et al., 2007; Feingersch and Béjà, 2009; Danhorn et al., 2012); and, finally, (v) if two rhodopsins with opposite activities are expressed from the same clone (that is, an outward proton transporter and an inward chloride transporter).
With all the above restrictions in mind, we suggest that our designed screening strategy is currently the best option for discovering novel microbial rhodopsins. With more libraries from different environments screened, and with automation of the method, we envision the finding of diverse rhodopsin groups with new transport activities possibly providing a molecular basis for developing novel optogenetic tools (Boyden et al., 2005; Zhang et al., 2007; Inoue et al., 2014).
Accession codes
References
Balashov SP, Imasheva ES, Boichenko VA, Anton J, Wang JM, Lanyi JK . (2005). Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science 309: 2061–2064.
Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K . (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8: 1263–1268.
Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP et al. (2000a). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289: 1902–1906.
Béjà O, Pinhassi J, Spudich JL (2013). Proteorhodopsins: widespread microbial light-driven proton pumps. In: Levin SA (ed). Encyclopedia of Biodiversity. Elsevier: New York, NY, USA, pp 280–285.
Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF . (2001). Proteorhodopsin phototrophy in the ocean. Nature 411: 786–789.
Béjà O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP et al. (2000b). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2: 516–529.
Danhorn T, Young CR, Delong EF . (2012). Comparison of large-insert, small-insert and pyrosequencing libraries for metagenomic analysis. ISME J 6: 2056–2066.
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469.
Dupont CL, Rusch DB, Yooseph S, Lombardo MJ, Richter RA, Valas R et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6: 1186–1199.
Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H . (2014). Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev 114: 126–163.
Feingersch R, Béjà O . (2009). Bias in assessments of marine SAR11 biodiversity in environmental fosmid and BAC libraries? ISME J 3: 1117–1119.
Garcia SL, McMahon KD, Martinez-Garcia M, Srivastava A, Sczyrba A, Stepanauskas R et al. (2013). Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J 7: 137–147.
Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F . (2013). Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci Rep 3: 2471.
Grote M, O'Malley MA . (2011). Enlightening the life sciences: the history of halobacterial and microbial rhodopsin research. FEMS Microbiol Rev 35: 1082–1099.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O . (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321.
Heberle J, Deupi X . (2014). Retinal proteins - you can teach an old dog new tricks. Biochim Biophys Acta 1837: 531–532.
Inoue K, Kato Y, Kandori H . (2014). Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol 23: 91–98.
Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K et al. (2013). A light-driven sodium ion pump in marine bacteria. Nat Commun 4: 1678.
Keffer JL, Hahn MW, Maresca JA . (2015). Characterization of an unconventional rhodopsin from the freshwater Actinobacterium Rhodoluna lacicola. J Bacteriol 197: 2704–2712.
Lanyi JK, Oesterhelt D . (1982). Identification of the retinal-binding protein in halorhodopsin. J Biol Chem 257: 2674–2677.
Lanyi JK, Weber HJ . (1980). Spectrophotometric identification of the pigment associated with light-driven primary sodium translocation in Halobacterium halobium. J Biol Chem 255: 243–250.
Martínez A, Bradley AS, Waldbauer J, Summons RE, DeLong EF . (2007). Proteorhodopsin photosystem gene expression enables photophosphorylation in heterologous host. Proc Natl Acad Sci USA 104: 5590–5595.
Oesterhelt D, Stoeckenius W . (1971). Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol 233: 149–152.
Philosof A, Béjà O . (2013). Bacterial, archaeal and viral-like rhodopsins from the Red Sea. Environ Microbiol Rep 5: 475–482.
Sabehi G, Loy A, Jung KH, Partha R, Spudich JL, Isaacson T et al. (2005). New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol 3: e173.
Sorek R, Zhu YW, Creevey CJ, Francino MP, Bork P, Rubin EM . (2007). Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318: 1449–1452.
Spudich JL, Yang CS, Jung KH, Spudich EN . (2000). Retinylidene proteins: structures and functions from archaea to humans. Annu Rev Cell Dev Biol 16: 365–392.
Whelan S, Goldman N . (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699.
Yoshizawa S, Kumagai Y, Kim H, Ogura Y, Hayashi T, Iwasaki W et al. (2014). Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc Natl Acad Sci USA 111: 6732–6737.
Yutin N, Koonin EV . (2012). Proteorhodopsin genes in giant viruses. Biol Direct 7: 34.
Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K . (2007). Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci 8: 577–581.
Acknowledgements
We thank Jason Poulos from Librede Inc. for assembling the LED device, Alon Philosof for his help with the genomic assemblies and Ed DeLong for kindly sending us fosmid HF10_19P19 for initiating this project. This work was supported by Israel Science Foundation grants 1769/12, the I-CORE Program of the Planning and Budgeting Committee and the Grand Technion Energy Program (GTEP), is part of The Leona M. and Harry B. Helmsley Charitable Trust reports on Alternative Energy series of the Technion—Israel Institute of Technology and the Weizmann Institute of Science, and is also supported by the Technion’s Lorry I. Lokey Interdisciplinary Center for Life Sciences and Engineering and the Russell Berrie Nanotechnology Institute. Fosmid sequences were deposited in GenBank under accession numbers KT201082−KT201092.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Pushkarev, A., Béjà, O. Functional metagenomic screen reveals new and diverse microbial rhodopsins. ISME J 10, 2331–2335 (2016). https://doi.org/10.1038/ismej.2016.7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.7
This article is cited by
-
Rhodopsin-mediated nutrient uptake by cultivated photoheterotrophic Verrucomicrobiota
The ISME Journal (2023)
-
Expression of Xanthorhodopsin in Escherichia coli
The Protein Journal (2023)
-
Heterologous expression and cell membrane localization of dinoflagellate opsins (rhodopsin proteins) in mammalian cells
Marine Life Science & Technology (2020)
-
A distinct abundant group of microbial rhodopsins discovered using functional metagenomics
Nature (2018)