Abstract
Plants associate through their roots with fungal assemblages that impact their abundance and productivity. Non-mycorrhizal endophytes constitute an important component of such fungal diversity, but their implication in ecosystem processes is little known. Using a selection of 128 root-endophytic strains, we defined functional groups based on their traits and plant interactions with potential to predict community assembly and symbiotic association processes. In vitro tests of the strains’ interactions with Arabidopsis thaliana, Microthlaspi erraticum and Hordeum vulgare showed a net negative effect of fungal colonization on plant growth. The effects partly depended on the phylogenetic affiliation of strains, but also varied considerably depending on the plant-strain combination. The variation was partly explained by fungal traits shared by different lineages, like growth rates or melanization. The origin of strains also affected their symbioses, with endophytes isolated from Microthlaspi spp. populations being more detrimental to M. erraticum than strains from other sources. Our findings suggest that plant–endophyte associations are subject to local processes of selection, in which particular combinations of symbionts are favored across landscapes. We also show that different common endophytic taxa have differential sets of traits found to affect interactions, hinting to a functional complementarity that can explain their frequent co-existence in natural communities.
Similar content being viewed by others
Introduction
Root endosymbiotic fungi have impacts on ecosystem functioning through their effects on plant productivity and community assembly (Bever et al., 2010, 2012). Some, such as mycorrhizal fungi, develop mutualistic interactions that allow plants to exploit habitats that would otherwise be inaccessible to them, and to boost their competitiveness over plants lacking these associations (Bever et al., 2010). Others have evolved pathogenic lifestyles and can reduce considerably their hosts’ fitness, thus contributing to the diversity of plant communities (Van der Putten et al., 1993; Wardle et al., 2004; Mangan et al., 2010). In addition to these relatively well-defined symbionts, healthy plant roots harbor a broad diversity of other fungi, referred to as root endophytes (Rodriguez et al., 2009; Sieber and Grünig, 2013). The effects of root endophytes on their hosts’ development are poorly known, and hence their function in natural ecosystems remains cryptic (Mandyam and Jumpponen, 2005).
Non-mycorrhizal endophytes represent the largest fraction of the fungal diversity within roots, and they are found in all plants and land ecosystems (Vandenkoornhuyse et al., 2002; Sieber and Grünig, 2013). They form polyphyletic ensembles seemingly adapted to the root environment, as their structure and composition differ from those in the neighboring soil and plant organs (Maciá-Vicente et al., 2012; Coleman-Derr et al., 2016). Because the endophyte concept constitutes a catchall classification encompassing all symbionts in the interior of healthy plant tissues (Rodriguez et al., 2009), it is likely to lump together fungal lineages with heterogeneous ecological roles. For example, it is argued that endophytes develop symbioses ranging from parasitic to mutualistic (Mandyam and Jumpponen, 2015) or that depend on the trade-off of particular resources (Newsham, 2011). Moreover, the occurrence of particular endophytes depends on host identity and environmental conditions (for example, Maciá-Vicente et al., 2008a, 2012; Wehner et al., 2014; Glynou et al., 2016), they can occupy different root compartments, or follow distinctive patterns of colonization (Maciá-Vicente et al., 2008b, 2009a, 2012; Peterson et al., 2008; Atsatt and Whiteside, 2014). A comprehensive characterization of the symbiotic roles played by endophytes is necessary to understand the evolutionary processes determining the plant-associated fungal diversity and its contribution to the feedbacks that sustain natural communities (Bever et al., 2012).
Many studies have aimed to assess the natural function of endophytes by reproducing their interaction with plants under controlled conditions (for example, Usuki and Narisawa, 2007; Maciá-Vicente et al., 2008b, 2009a,b; Tellenbach et al., 2011; Keim et al., 2014; Mandyam and Jumpponen, 2014, 2015). They provide examples of specific associations between particular fungal and plant genotypes, but they are difficult to extrapolate to general scenarios owing to a high intra-specific variability of the interactions (Tellenbach et al., 2011; Mayerhofer et al., 2012) and to the difficulty in detecting responses in either symbiont (Mandyam and Jumpponen, 2005). Alternative approaches based on the measurement of fungal traits have been proposed to unravel the implication of fungi in ecosystem dynamics (Aguilar-Trigueros et al., 2014, 2015). Classifications of species based on their sets of traits have been used to define major life history strategies, which, in turn, can predict patterns of biodiversity, community assembly and natural associations (Chagnon et al., 2013).
Trait-based approaches have proven valuable to identify relationships between life history and functional traits of arbuscular mycorrhizal fungi and their plant interactions (for exampl, Powell et al., 2009; Maherali and Klironomos, 2012; Chagnon et al., 2013). For example, differences across arbuscular mycorrhizal fungal lineages in rates of nutrient exchange with hosts, sporulation and biomass allocation to mycelial compartments, are linked with their association and interaction with particular plants, their biogeographic and successional patterns, and their community structure (Chagnon et al., 2013). Distinctive traits have also been used to define groups of non-mycorrhizal endophytes, like in the so-called dark-septate endophytes (DSE; Jumpponen and Trappe, 1998). But how these traits are relevant for the symbiosis is seldom known, and systematic studies on the patterns of distribution and evolution of characters across endophytic lineages are lacking (Aguilar-Trigueros et al., 2014).
In this study, we examine the influence of phylogeny and traits of root-endophytic fungi on their interaction with plants. We employ a collection of strains isolated from different plant species, geographical locations and habitats. Most of them originate from a screening of the non-mycorrhizal plant Microthlaspi spp. (Brassicaceae) across Europe (Glynou et al., 2016), which harbored a broad diversity of endophytes. In it, a few endophytes with disparate phylogenetic affiliations, like Fusarium spp., Alternaria spp. and Cadophora spp., were ubiquitous and co-existed frequently in the same root communities, but displayed distinctive distribution patterns and ecological preferences. Therefore, our collection provides a basis to assess patterns of trait variation across fungal lineages, geography and ecological conditions. Here, we measure life history traits of endophytes, such as growth rates and sporulation capacity, as well as traits proposed to be potentially functional for the symbiosis, like hyphal melanization and production of intraradical microsclerotia—defining characters of DSE—and enzymatic activities that can facilitate host nutrient uptake or assist fungal penetration of plant tissues (Mandyam et al., 2010). In addition, we assess the effect of strains on the growth of Microthlaspi erraticum, its confamilial Arabidopsis thaliana and the gramineous Hordeum vulgare (Poaceae). Our aim is to test how the interactions between root-endophytic fungi and plants are influenced by phylogenetic conservatism, as well as by convergent traits and ecological origins of fungi that are unrelated to phylogeny.
Materials and methods
Fungal strains and plant material
One hundred and twenty-eight fungal strains isolated from roots of different plant species and geographical locations were used in this study. The majority originate from Microthlaspi spp. (Glynou et al., 2016), whereas others were isolated from Salicornia spp. (Amaranthaceae). Endophytes were isolated in culture after the surface-sterilization of roots as described by Maciá-Vicente et al. (2012), and selected prior to their identification by choosing morphologically divergent strains from different plants/locations. In addition, we obtained Serendipita indica (syn: Piriformospora indica) CBS 125645 from the KNAW-CBS Fungal Biodiversity Centre. S. indica has been thoroughly studied as a model endophyte with a mutualistic interaction with multiple plants (Banhara et al., 2015). A description of all strains is provided in Supplementary Table S1.
The plants A. thaliana ecotype Col-0, M. erraticum and H. vulgare cv. Barke (barley) were used as hosts in plant–endophyte interaction assays. Seeds of A. thaliana were provided by the Laboratory of Plant Physiology of Wageningen University. Seeds of M. erraticum were collected from a field population in Germany (Mp_K11; Ali et al., 2016). Barley seeds were provided by the company Saatzucht Josef Breun GmbH & Co. KG (Herzogenaurach, Germany).
Molecular characterization of strains
We obtained the sequences of the ribosomal DNA internal transcribed spacer regions (ITS) of all strains. ITS sequences from most strains were already available from Glynou et al. (2016), and the rest were obtained as described therein. We also followed the procedures in Glynou et al. (2016) to assign the strains to taxa and to group them into operational taxonomic units (OTUs). In brief, genomic DNA was extracted from fungal mycelia using the BioSprint 96 DNA Plant Kit (Qiagen, Hilden, Germany) on a KingFisher Flex 96 robotic workstation (Thermo Fisher Scientific, Waltham, MA, USA). ITS sequences were amplified and sequenced using the primer pair ITS1F/ITS4 (White et al., 1990; Gardes and Bruns, 1993), and they were then classified at different taxonomic precisions with the Naive Bayesian classifier tool of Mothur v1.34.4 (Wang et al. 2007; Schloss et al. 2009), based on comparisons with the UNITE database of curated fungal ITS sequences (Kõljalg et al., 2013). Strains were grouped into OTUs according to ITS pairwise similarity of at least 97%, using the BLASTClust program (Altschul et al., 1997). The taxonomic classification of strains and the GenBank accession numbers of all sequences are provided in Supplementary Table S1.
We built a molecular phylogeny with the ITS sequences using Bayesian inference. The ITS1, 5.8S and ITS2 regions were independently aligned using MAFFT v7.123b (Katoh and Standley, 2013), and ambiguously aligned regions were removed using Gblocks v0.91b (Castresana, 2000). Two parallel MCMC analyses, using the GTRGAMMA model with independent parameter estimates for each partition, were run in MrBayes v3.2.2 (Huelsenbeck and Ronquist, 2001) for 10 M generations with sampling every 100th generation and 30% burn-in. An ultrametric majority-rule consensus tree was used in subsequent analyses. Whereas the ITS regions are not suitable for phylogenies involving distantly related taxa owing to their variability, our trimmed alignment consisted mostly of the conserved 5.8S gene. The latter has been used in phylogenies of highly divergent taxa (Redecker et al., 1999), and our resulting tree reflected the OTU relationships among strains (Supplementary Figure S2).
Morphological and physiological characterization of strains
The strains were maintained in triplicate cultures on corn meal agar (CMA, Sigma-Aldrich, St. Louis, MO, USA) and malt extract agar (MEA, Applichem, Darmstadt, Germany). We recorded the presence/absence of conidia and darkly pigmented (dematiaceous) mycelia in cultures for up to 3 months. Radial growth rates were measured three days after plating on each medium and reported as millimeters of colony expansion per day. We measured the production of extracellular enzymes using custom plate assays. Cellulase activity was assessed by the clearing halo produced by 7-day-old colonies on Czapek-Dox agar with 0.5% (w/v) carboxymethylcellulose sodium salt as sole carbon source (Johnsen and Krause, 2014). Peptidase, pectinase, laccase and peroxidase activities were measured following the methods described by Basiewicz et al. (2012). In addition, the ability of strains to solubilize mineral phosphate was measured as in Zavala-Gonzalez et al. (2015). Cellulase, pectinase and phosphate solubilization activities were measured as the proportional width of the clearing halo respect to colony diameter. Peptidase, laccase and peroxidase activities were rated according to a 1–4 semi-quantitative scale.
Arabidopsis and microthlaspi inoculation assays
We tested the effect of colonization by individual strains on the development of A. thaliana and M. erraticum using an in vitro assay (Supplementary Figure S3). Surface-sterilized seeds were plated on half-strength Murashige-Skoog basal salt solid medium (MS, Sigma-Aldrich; Murashige and Skoog, 1962), stratified for 2 days at 4 °C in the dark, and then incubated for 7 days at 23 °C under continuous illumination (80 μmol m−1 s−1). Upon emergence of the first true leaves, seedlings were transferred to 24-well plates containing MS medium and maintained in the same incubation conditions. After 10 days, plants were inoculated with individual strains or left un-inoculated in controls. Because many strains did not sporulate in culture, inoculation was performed by puncturing the margin of actively growing colonies on CMA with a sterilized toothpick to collect a small amount of mycelium, and then transferring it a few millimeters from the crown of plants by inserting it in the agar. Ten days after inoculation, the development of mycelium in roots was confirmed under a stereomicroscope. Symptoms of chlorosis and/or necrosis in leaves were rated on a semi-quantitative scale (0=none, 1=up to 30% chlorotic/necrotic leaves, 2=30–60%, 3=>60%, 4=dead plants), and the fresh weight of the aerial tissues was measured. Every treatment consisted of five replicates, performed simultaneously in a separate 24-well plate each. The layout of treatments was randomized within wells to minimize potential effects on the data owing to position. Experiments were performed in batches including 23 strains and a control treatment each, and measurements for each fungal treatment were compared only to its respective control. To assess the reproducibility of assays, we repeated them for 34 strains in A. thaliana (Supplementary Figure S4).
Barley inoculation assays
The effects of root colonization on barley were assessed using a standard in vitro assay (Dufresne and Osbourn, 2001; Maciá-Vicente et al., 2008b). In brief, 2-day-old seedlings obtained from surface-sterilized seeds were planted in glass tubes with 30 ml sterilized, hydrated vermiculite. Four 5-mm-diameter plugs taken from the margin of actively growing colonies on CMA were used as inoculum, by placing them 2–3 cm deep in the vermiculite. Control tubes were mock-inoculated with sterile CMA plugs. Plants were grown under long day conditions (16 h:8 h, light:dark, 80 μmol m−1 s−1) at 23 °C. The fresh weight of roots and shoots was measured after 10 days. In this case, we did not score symptoms because they were seldom evident in leaves, and detrimental effects were manifested by reductions on biomass (Supplementary Figure S3). Treatments consisted of 10 replicates, which were performed in batches of three to nine strains and one un-inoculated control treatment each. We repeated these assays for five strains to test their reproducibility (Supplementary Figure S4).
We assessed the endophytic colonization of roots by cultivation and microscopy methods. In the first case, one to two roots per plant were surface-sterilized for 1 min with 0.5% sodium hypochlorite, rinsed with sterilized deionized water, and cut into 0.5 cm pieces. Ten root pieces per plant were randomly selected, dry-blotted onto sterilized paper and plated on CMA. The efficacy of the sterilization procedure was assessed in a subset of 30 root pieces per treatment with the imprint method (Hallmann et al., 2006). The percentage of root pieces colonized was recorded 5–7 days later. For the microscopical observation of root colonization, we randomly selected three barley plants per treatment. One entire seminal root per plant was cleared overnight in a 1 m KOH solution, stained with acidified lactophenol blue and kept in acidified glycerol until observation. Samples were observed in squash preparations, in which epiphytic and/or endophytic root colonization, and the presence of microsclerotia were recorded (Supplementary Figure S5). The latter was considered as a fungal trait in subsequent analyses.
Statistical analyses
Data organization
Statistical analyses were performed using R v3.0.2 (R Core Team, 2013). Data from the inoculation assays were first assessed for normality and homoscedasticity, and then treatments were compared using analysis of variance or the Kruskal–Wallis test. Subsequent pairwise comparisons of each fungal treatment against its respective control were done by either t-tests or Wilcoxon tests with a Holm–Bonferroni correction. In order to incorporate these data into further analyses, we calculated the effect size of biomass variables from each treatment respect to its un-inoculated control. Effect sizes with 95% confidence intervals were calculated according to the Cohend’s d statistic (Cohen, 1988) using function cohen.d in package effsize v0.5.4 (Torchiano, 2015), which measures the difference in means and standardizes it by their pooled s.d.
Fungal identifications resulted in several strains isolated from the same plant population being assigned to the same OTU. We considered them likely to belong to the same genets. In order to avoid repeated observations that could inflate the significance of tests, we thinned our datasets to 115 strains representing unique potential genets (Supplementary Figure S2). Pairs of strains within each potential genet often showed similar effects on plant growth (Supplementary Figure S6), and hence alternative selections of strains hardly changed results in downstream analyses.
Analysis of fungal traits
We used principal component analysis with standardized morphological and physiological trait data to summarize the differences among the 115 selected strains. We assessed the goodness of fit of the principal component analysis using the broken-stick criterion, which tests the cumulative percentage of variance explained respect to a random breakdown of variance. Individual variables significantly contributing to axes were identified using the equilibrium circle method (Legendre and Legendre, 2012).
Measurement of phylogenetic signal
We calculated the phylogenetic signal in the response of plant growth to fungal inoculation with the K statistic (Blomberg et al., 2003), using function phylosig in package phytools v0.5 (Revell, 2011). The method is used to assess conservation of traits among species, with K=0 indicating absence of a phylogenetic signal, and K<1 or K>1 resemblance lower or higher than expected under Brownian motion evolution. Because simultaneous inferences of phylogenetic signal between species and within species are difficult to interpret (Blomberg et al., 2003), we only included in these analyses individual values for each OTU as the mean of the effects by its strains. Sampling errors of within-OTU variability were incorporated following Ives et al. (2007), assuming variances for OTUs with only one strain equal to the mean of the overall within-OTU variance. The significance of K was assessed by comparing with a random shuffle of values at the tips of the phylogenetic tree. We also tested for phylogenetic signal in the interactions of strains within the orders Pleosporales, Hypocreales or Helotiales, separately.
Contribution of strain features to plant interactions
To assess the influence of phylogeny, traits and origin of strains on plant growth, we applied the variation partitioning method described by Desdevises et al. (2003) with function varpart in package vegan v2.2–1 (Oksanen et al., 2015). We performed the tests independently for each plant species, including as a response variable the effect size of their biomass as affected by every fungus. The variation was decomposed into three independent explanatory matrices gathering variables related to the strains’ traits, origin (geographical coordinates, and natural host as Microthlaspi spp. or others) and phylogeny (principal coordinates (PCs) obtained from the phylogenetic tree). We retained only 19 PCs that were significantly correlated in linear regressions (P<0.05) with the effects on at least one plant, representing a combination of early (PCs 1 and 2) and late phylogenetic divergences. The significance of the variance fractions explained by each component was tested using permutation tests with pseudo F-ratios.
To estimate the contribution of each fungal lineage to tree-wide variation in traits, we calculated their contribution indices (Moles et al., 2005) using the aotf function in the program phylocom v4.2 (Webb et al., 2008). The index measures the proportional contribution of individual nodal divergences along the phylogeny to extant trait variation. Statistical support is assessed by comparing the values with those obtained by a random shuffle of traits at the tree tips. A trait can be considered conserved if more variation is explained by ancient than by recent divergences (Maherali and Klironomos, 2012).
We tested for potential relationships between individual trait/origin variables and the effects on plant biomass, using phylogenetic generalized least squares to account for phylogenetic signal in the data. Phylogenetic generalized least squares estimates regression parameters weighted by phylogenetic signal measured as Pagel’s λ (with 0 and 1 indicating no or strong signal, respectively; Pagel, 1999), and it is equivalent to an ordinary least squares model when the signal is absent in the residuals (Symonds and Blomberg, 2014). These analyses were carried out using function pgls in package caper v0.5.2 (Orme et al., 2011).
Results
Taxonomic classification of strains
The strains were classified in 54 OTUs and ascribed to 17 families in 11 orders. Among the 115 strains representing likely independent genets, 111 (96.5%) were species of Ascomycota and four (3.5%) of Basidiomycota. The most frequently encountered orders were Pleosporales and Hypocreales, with 56 (48.7%) and 39 (34%) strains belonging to 26 (48.1%) and 11 (20.4%) OTUs, respectively (Supplementary Figure S1). They were followed by Helotiales, with 10 strains (8.7%) in seven OTUs (13%), whereas other orders were represented by two or less strains each (Supplementary Figure S1). Within Pleosporales, 29 strains (25.2%) in seven OTUs (13%) belonged to the family Pleosporaceae and were mainly represented by OTUs related to Alternaria spp. Other OTUs within Pleosporales were designated as family incertae sedis or remain unclassified. Most members of Hypocreales belonged to Nectriaceae, with 31 strains (27%) out of which 29 belonged to six OTUs classified as Fusarium spp. The most frequent of these were OTU001 with affinities to Fusarium tricintum and Fusarium avenaceum, and OTU003 related to Fusarium oxysporum. Other species of Hypocreales were assigned to Emericellopsis and Ilyonectria, with families incertae sedis. Six Helotiales strains (5.2%) had ITS affinities with the genus Cadophora with family incertae sedis, and are referred to as Cadophora-like onwards (Supplementary Figure S1).
Characterization of strain traits
We measured variables of morphology, growth rates and enzymatic and phosphate solubilization activities in all fungal strains (Supplementary Table S1; Supplementary Figure S5). A principal component analysis ordination of the trait data explained an overall 48.4% of the variance (Figure 1), larger than that explained by a random breakdown of variance (36%). The first component collected most information related to the strains’ growth rates and clearing halos (Figure 1a). The second component mainly represented variation in peroxidase activity, pigmentation and production of intraradical microsclerotia (Figure 1a). The ordination of strains reflected their phylogenetic affinities (Figure 1b), with a clear separation of Hypocreales, Pleosporales and Cadophora-like strains along the first axis. Members of Nectriaceae formed a compact cluster clearly separated from other Hypocreales and most other fungi (Figure 1b). Strains within Pleosporaceae showed a tendency toward a high peroxidase activity and the formation of dematiaceous mycelia and microsclerotia, although they also showed a wide variability in these characters (Figure 1b).
Principal component analysis (PCA) ordination of fungal endophytic strains according to their physiological and morphological traits. The two axes represent 36.2% and 12.2% of the data variance, respectively. (a) PCA scores showing the contribution of each trait to the separation of the strains, as indicated by the direction and magnitude of the respective arrows. Variables in bold contributed significantly to the variance, according to the equilibrium circle method. (b) Ordination of strains according to their traits. Strains belonging to the three most represented fungal orders are shown in different colors (see color key). Different symbols within each of these orders indicate strains belonging to different families, or to paraphyletic groups at that taxonomic level. Ellipses delimit 95% confidence intervals around the strains of Nectriaceae (solid circles), Pleosporaceae (solid squares) and Helotiales incertae sedis (=Cadophora-like, solid triangles). Abbreviations: cel.g, growth rate on cellulose; cel.h, degradation halo on cellulose; cel.i, cellulase activity; cma, growth rate on CMA; dem, pigmentation; gel.g, growth rate on gelatin; lacc.i, laccase activity; mea, growth rate on MEA; mic, production of microsclerotia; P, phosphorus solubilization; pec.g, growth rate on pectin; pec.h, degradation halo on pectin; pec.i, pectinase activity; per.i, peroxidase activity; prot.i, protease activity; spo, production of conidia.
Effect of fungal strains on plant growth
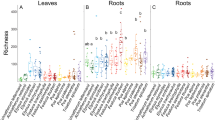
Inoculation assays of individual strains in A. thaliana, M. erraticum and H. vulgare yielded a wide range of growth responses, ranging from a strong inhibition to a moderate stimulation of plant biomass production in comparison to un-inoculated controls (Figure 2; Supplementary Figure S3). The overall effect of fungal inoculation was negative for all host species (W=395–1685, P<0.001), but it was less marked in barley. Similar results were obtained when considering variables of plant development other than total biomass, because they were strongly collinear with it (Pearson’s r<−0.74, P<0.001 for symptoms data in both Brassicaceae; r>0.97, P<0.001 for the effects on shoot and root biomass in H. vulgare). Moreover, similar effects were observed in repetitions of these experiments with subsets of strains (Supplementary Figure S4).
Interactions between 128 fungal endophytic strains and the plants Arabidopsis thaliana, Microthlaspi erraticum and Hordeum vulgare. Selected fungal traits are also shown. Bars represent effect sizes (Cohend’s d)±95% confidence intervals for the interactions with plants, and mean values±s.e. for the growth rates of strains on corn meal agar (CMA) and barley root colonization. Boxplots at the bottom of graphs represent the overall data distribution for each variable. Points next to bars for A. thaliana and M. erraticum indicate average scores >1 in a semi-quantitative scale of symptoms. Solid and empty bullets in the qualitative traits of dematiaceous mycelium (Demat.), production of conidia, and production of microsclerotia (Microscl.) indicate presence or absence of the character, respectively.
We only found conservatism in the response of M. erraticum to fungal inoculation (Table 1). OTUs within the Hypocreales had a conserved effect on A. thaliana but non-significant signals in their interactions with M. erraticum and H. vulgare (Table 1). The responses to fungal inoculation varied considerably across plants and fungal lineages. M. erraticum and H. vulgare were most negatively affected by Fusarium spp. strains, whereas the strongest negative effects on growth of A. thaliana were caused by members of the Pleosporaceae (Figure 2, Supplementary Figure S7). Fungal OTUs with the strongest overall virulence towards either plant species, such as Fusarium spp. OTU001 and OTU003, and Alternaria sp. OTU008, also showed a broad within-group variability that spanned the entire range of interactions (Figure 2, Supplementary Figure S7). The effects of Cadophora-like strains were always close to neutrality (Figure 2, Supplementary Figure S7).
Fungal colonization of roots
Fungal root colonization was detected in most plants at the moment of sampling. In H. vulgare, we quantified the degree of colonization in culture, and we often observed it directly by light microscopy (Supplementary Figure S5). We did not find a significant phylogenetic conservatism in the fungal colonization of barley roots (K=0.7, P=0.9). Root colonization was negatively correlated, after controlling for phylogenetic signal, with the effect on total plant biomass of each strain (slope=−0.023±0.003, adj. R2=0.27, F124=32.82, P<0.001, λ=0). A similar result was obtained for the effect on shoot and root biomass (Supplementary Table S2). Moreover, root colonization was positively correlated with all variables of fungal growth (P<0.01).
Contribution of strain features to plant interactions
We evaluated the contribution of strain variables related to phylogeny, traits and origins, to the effect of fungal inoculation on plant growth (Figure 3). Models for each plant explained a significant proportion of the variation in their growth response to fungal inoculation (48–59%, P<0.001). The phylogeny of strains predicted the largest fraction of the variance in A. thaliana and M. erraticum (45.1 and 35.7%), but it was less informative than the strains’ traits in H. vulgare (25.1% respect to 33%). Some fungal clades contributed greatly to the overall phylogenetic signal, mainly representing late divergences in the phylogenetic tree at the OTU level (Figure 4). Fusarium sp. OTU001 had the largest contribution to overall variance in the responses of M. erraticum and H. vulgare, whereas divergences in growth of A. thaliana were most affected by several pleosporaceous OTUs (Figure 4). These late divergences contrast with those obtained for mycelial traits, which tended to be greater earlier in the phylogeny (Figure 4, Supplementary Figure S8). The response of both Brassicaceae had little dependence on the strains’ traits alone (4–8%), which had an effect partly indistinguishable from that of the strains’ phylogeny in M. erraticum (Figure 3). Fungal traits explained a significant amount of the interactions with H. vulgare (33.1%). Only the response of M. erraticum to colonization was significantly correlated with the strains’ origin (Figure 3).
Euler diagrams of variation partitioning analysis, showing the effects of traits, origin and phylogeny of the endophytic strains on the growth of Arabidopsis thaliana, Microthlaspi erraticum and Hordeum vulgare. Values indicate the proportion of the variation explained (adjusted R2) by each fraction, corresponding to the pure effects of explanatory variables, or their shared effects (overlapping fractions). Values in bold are significant (P<0.05). Zero and negative values are not shown.
Nodal contributions to tree-wide variation in the response of Arabidopsis thaliana, Microthlaspi erraticum and Hordeum vulgare to fungal inoculation, and to selected fungal traits relative to their morphology and physiology. Circles in the tree nodes indicate contribution indices that are significant (P<0.05), as compared with a distribution of 999 values calculated by a random shuffle of trait values across the tips of the phylogeny. Shading of circles represents the relative contribution of individual nodes to extant trait variation (see key). Scatterplots in insets show the relationship between the contribution indices and the respective age of nodes, with black and white circles indicating contribution indices that are significant or not, respectively.
Phylogeny-independent determinants of plant–endophyte interactions
We assessed the correlation between individual strain variables included in the variation partition and the response of plants to fungal colonization using phylogenetic generalized least squares to subtract phylogenetic signal. All descriptors of hyphal growth were strongly associated with negative effects on the development of the three plants (Table 2). Of the physiological characteristics of strains, only laccase and pectinase activities showed a significant association with biomass of H. vulgare (Table 2). Production of dematiaceous mycelium and conidia had contrasting positive and negative relationships with the development of individual plant species, respectively (Table 2). Interestingly, strains originally isolated from Microthlaspi spp. showed a stronger virulence than strains from other sources toward the congeneric M. erraticum (Table 2).
Discussion
We provide evidence that the effects of non-mycorrhizal fungal root endophytes on plant growth are strongly influenced by the phylogeny of fungi. However, the phylogenetic signal is mostly explained by recent divergences that indicate little conservatism in the evolution of interactions. Moreover, particular fungal traits shared by phylogenetically dispersed taxa affected to a different extent the plant responses to fungal inoculation. These effects always followed a similar trend in different plant species, suggesting a direct relation of the traits with specific types of associations, or their linkage with other characters relevant to the symbiosis (Treseder and Lennon, 2015). The collation of these traits across strains allows a rough functional classification of the fungal diversity included in our study, and to hypothesize about their influence in the assembly of natural root-endophytic communities.
Effect of fungal colonization on plant development
The net effect of fungal colonization on plant biomass was negative, consistent with previous results based on the controlled inoculation of plants with root endophytes (Tellenbach et al., 2011; Mayerhofer et al., 2012; Keim et al., 2014; Mandyam and Jumpponen, 2015). Our experimental system included the plant as the sole carbon source to sustain fungal growth, which conditioned a strong negative correlation between fungal development and plant biomass, similar to that reported in other endophytic interactions (Tellenbach et al., 2011). However, the negative responses were often small, and strong compromises of plant growth and development of symptoms were scarce. Most root endophytes do not seem an important burden to their hosts, suggesting that their parasitism may be easily compensated by slight enhancements of plant fitness in their natural habitat. Mutualistic interactions depend on a balance between net costs and benefits provided by symbionts, whereby they can become parasitic in the absence of the ecological factors that drive the relationship. There are multiple instances of non-mycorrhizal endophytes providing their hosts with benefits when exposed to external factors, such as pathogens (Maciá-Vicente et al., 2008b), environmental stress (Rodriguez et al., 2008) or nutrient shortages (Usuki and Narisawa, 2007; Behie et al., 2012; Hiruma et al., 2016). Endophytes are likely implicated in different yet unknown context-dependent trade-offs associated with conditions not reproduced in our system. A blind testing of multiple environmental factors was out of the scope of our study, but further work in this direction may help to unravel context-dependent symbioses.
Phylogenetic conservatism of plant–endophyte interactions
Plant responses to endophytic colonization partly depended on the phylogenetic relations of strains, suggesting the evolution of distinctive strategies for the interaction with hosts. These effects varied markedly across plant species, in line with studies evidencing a large dependency of plant–fungus symbioses on the specific combination of partners (Klironomos, 2003; Mandyam and Jumpponen, 2015). Much of the variation was associated with negative feedbacks with fungi related to well-known pathogens, particularly species within Fusarium, and Pleosporaceae like Alternaria spp. These lineages often dominate roots of healthy wild plants (Maciá-Vicente et al., 2008a, 2012; Knapp et al., 2012; Sánchez Márquez et al., 2012; Glynou et al., 2016), where their detrimental effects appear to be mitigated by environmental conditions and/or interactions with extant microorganisms. But the linkage between particular responses and well-defined fungal clades was not clear-cut, because several strains in the most virulent groups had little impact on plant growth. It would be reasonable to expect a clear differentiation in the associations involving particular fungal lineages, given the diverse life histories that determine distinctive physiological and morphological adaptations. Such trends have been identified in endophytes (Mayerhofer et al., 2012), but they are generally diffuse owing to a high intra-specific variability that often exceeds the variation between species (Tellenbach, et al. 2011; Mandyam and Jumpponen, 2014). Fungal determinants of the symbiosis might be subject to rapid and recent change over evolutionary time, perhaps separating different genetic populations within species. For example, Fusarium species show a wide intra-specific variability in their virulence because they have pathogenicity genes subject to strong diversifying pressure or that can be horizontally transferred with mobile chromosomes (Ma et al., 2010; Sperschneider et al., 2015). Further, Cheikh-Ali et al. (2015)) found that root endophytes of a same phylotype isolated from distant localities expressed divergent morphological and physiological characters. The high intra-specific variation of plant–endophyte interactions has likely ecological implications for the local assembly of natural communities, because it might be a consequence of adaptations to local conditions. This would promote selection mosaics across landscapes in which particular combinations of symbionts are favored by their joint response to extant conditions (Thompson, 2005; Piculell et al., 2008).
The broad variability in plant–endophyte interactions contrasts with patterns of evolution in functional traits of arbuscular mycorrhizal fungi, which appear to be phylogenetically conserved at the family level (Powell et al., 2009; Chagnon et al., 2013). The opposing patterns between mycorrhizal and non-mycorrhizal fungal endosymbionts probably reflect large differences in their specialization for the symbiosis. Unlike mycorrhizas, most endophytes are not bound to their plant hosts and can be found as saprotrophs in other substrata. Therefore, their evolution might be less subjected to constraints imposed by the symbiotic lifestyle.
Phylogeny-independent determinants of plant–endophyte interactions
The growth responses to fungal colonization varied among plant species. Interactions involving either brassicaceous host had a great dependency on the phylogenetic relations of strains, whereas in H. vulgare convergent fungal traits were more important in explaining the growth responses to inoculation. The distinctive results across plants could have been determined by methodological differences in the bioassays. Nevertheless, A. thaliana and M. erraticum were tested using a similar setup yet they had divergent responses to particular fungal groups like fusaria or Pleosporaceae strains, whereas growth patterns of M. erraticum had commonalities with those of barley. Besides, only M. erraticum showed a significant degree of conservatism in its response to close fungal relatives, which can be indicative of mutual adaptations between partners, given the affiliation of the plant with the natural host of most strains. Therefore, it seems likely that the variation in individual plant–endophyte combinations largely reflects actual specificities across partners.
In all cases, a significant amount of the variation was explained by traits shared by dispersed fungal clades, which were correlated with the outcome of interactions in a similar manner. Among these, the growth rates of strains were strongly associated with reductions of plant biomass, probably owing to the tendency of these strains to colonize host tissues systemically and to their larger demands on plant carbon. Likewise, strains capable to sporulate in culture tended to be more detrimental to plants, perhaps owing to the linkage of this trait with fast rates of mycelial growth.
Dematiaceous fungi were less prone to develop detrimental symbioses. Melanized hyphae, in combination with lack of spores in culture and the production of intraradical microsclerotia are defining attributes of DSE. These form a polyphyletic group of fungi frequently regarded as potential mutualists based on their high prevalence and ubiquity in roots (Mandyam and Jumpponen, 2005), although their symbiotic function is still elusive. Newsham (2011) detected a net positive effect of DSE on plant performance, associated with increments in nutrients uptake in the presence of soil organic matter. The hydrolytic capabilities of several DSE have been previously described and suggest that they are able to access detrital nutrient pools as saprotrophs (Caldwell et al., 2000; Mandyam and Jumpponen, 2005; Mandyam et al., 2010). Our inoculation assays did not include organic sources of nutrients available for the fungi other than the plant, but the saprotrophic capabilities of DSE might entail in nature a fitness benefit to hosts that could easily overcome their weak parasitism.
We were unable to detect direct substantial effects of the strains’ physiological activities on the outcome of interactions. Laccase and pectinase activities were the only traits somewhat associated with plant performance in barley. The expression of hydrolytic activities was highly variable in our assays, perhaps reflecting different substrate specificities and inducing conditions (Basiewicz et al., 2012), or unspecificities in the detection of particular activities (Johnsen and Krause, 2014). But this variation also highlights large differences among and within fungal taxa that suggest a broad diversity of potential interactions in response to the availability of substrates. This could be ultimately confirmed by comparing genomic traits relative to these activities and the assessment of their expression in planta (Lahrmann et al., 2015).
The original host of strains had a strong impact on their interactions with M. erraticum, in which strains isolated from congeneric plants were more virulent. This could indicate a certain host specificity of these strains that is backed by their phylogenetically conserved effect on this plant. Similar effects have been described for other root-endophytic (Tellenbach et al., 2011), pathogenic (Sacristán and García-Arenal, 2008) and mycorrhizal symbioses (Klironomos, 2003; Hoeksema and Thompson, 2007), what supports the hypothesis of symbiotic partners co-evolving in response to each other. Interestingly, stronger adaptations of root endophytes to their hosts often lead to an increased virulence (Tellenbach et al., 2011), as opposed to those involving mutualistic mycorrhizas (Hoeksema and Thompson, 2007). This hints to parasitism as the main lifestyle adopted by many root endophytes in nature.
Trait-based classification of strains
The grouping of strains based on the similarity of their traits clearly separated endophytic lineages that frequently dominate and co-exist in roots, particularly those related to Fusarium, Pleosporaceae and Cadophora-like (Glynou et al., 2016). The clustering was influenced by life history traits associated with the plant’s response to infection, suggesting a differential niche occupancy by groups of endophytes likely to condition their spatial distribution (Violle et al., 2007; Violle and Jiang, 2009). Fusarium spp. clearly differed from other taxa by fast growth and production of conidia. These characters are associated with an efficient ability of dispersal and resource colonization, which is consistent with the broad distribution across Europe observed for OTUs in this group (Glynou et al., 2016). Pleosporaceous and Cadophora-like strains, on the other hand, had slower growth rates and exhibited traits typical of DSE, and their geographical distribution apparently is constrained by environmental factors such as climatic and soil variables (Glynou et al., 2016). Spatial distribution is often used as a proxy of niche breadth, but this principle has been shown to be less applicable to microbes than to macroorganisms (Carbonero et al., 2014). Microorganisms highly specialized for a particular factor can have broad distributions if the latter is widespread, because they are less affected by other conditions than other generalist species. Consequently, the wide spatial occurrence of Fusarium spp. independently of other environmental factors could be a result of their efficient adaptations to colonize roots.
Fungal traits as determinants of community assembly
Our selection of strains represents well the composition and structure of endophytic assemblages associated with Microthlaspi spp. and other plants (Maciá-Vicente et al., 2008a, 2012; Sieber and Grünig, 2013; Keim et al., 2014), which are often co-dominated by species related to Fusarium, Pleosporaceae and Cadophora. The distinctive traits of these lineages are suggestive of different niche occupancies, therefore it is possible to associate the phylogenetic diversity of endophytic communities with processes of competition or complementarity among species. The co-occurrence of endophytes not sharing functional characteristics is indicative that competition is a main driver of community assembly, because functional complementarity reduces competition and promotes co-existence (Maherali and Klironomos, 2007, 2012). Conversely, communities shaped by environmental filtering usually show phylogenetic clustering of species with similar traits selected by the limiting factors. This could explain the low diversity in root-endophytic communities subject to salt stress, where pleosporaceous endophytes become enriched while otherwise dominant fusaria are absent (Maciá-Vicente et al., 2008a, 2012). Although functional complementarity among species enhances ecosystem function, the trait similarity of phylogenetically related endophytes (for example, different Fusarium spp. co-occurring in the same root) can lead to functional redundancy. This has shown to provide stability to plant–endophyte symbioses, because it prevents the loss of symbiotic functions with the replacement of fungal species across environmental gradients (Maherali and Klironomos, 2007).
Conclusions
Non-mycorrhizal fungal endophytes are pervasive in roots, hence they are likely to affect plant abundance and productivity in natural communities. Although our experimental system was artificial, it was adequate to address our objective to test interactions between plants and a large number of endophytic species. Under these conditions most endophytes behaved as weak parasites, but their performance varied across plant species and fungal taxa. Diverging endophyte lineages have evolved distinct strategies of plant symbiosis, but their associations were often variable, suggesting that they are subject to local processes of selection. Part of the variation in the interactions was explained by convergent fungal traits that differentiate categories of endophytes with potentially distinct niches. The functional complementarity of strains belonging to different groups is predicted by the structure of natural root-endophytic communities. The characterization of the endophytic diversity into potential functional groups will aid in the testing of further questions about their role in ecosystems. In particular, the assessment of the responses of plant–endophyte interactions to (a)biotic factors, including combinations of endophytes with different degrees of trait similarity and shared evolutionary history, will help unravel context-dependent symbioses adaptive under natural conditions.
References
Aguilar-Trigueros CA, Hempel S, Powell JR, Anderson IC, Antonovics J, Bergmann J et al. (2015). Branching out: towards a trait-based understanding of fungal ecology. Fungal Biol Rev 29: 34–41.
Aguilar-Trigueros CA, Powell JR, Anderson IC, Antonovics J, Rillig MC . (2014). Ecological understanding of root-infecting fungi using trait-based approaches. Trends Plant Sci 19: 432–438.
Ali T, Schmuker A, Runge F, Solovyeva I, Nigrelli L, Paule J et al. (2016). Morphology, phylogeny, and taxonomy of Microthlaspi (Brassicaceae: Coluteocarpeae) and related genera. Taxon 65: 79–98.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Atsatt PR, Whiteside MD . (2014). Novel symbiotic protoplasts formed by endophytic fungi explain their hidden existence, lifestyle switching, and diversity within the plant kingdom. PLoS One 9: e95266.
Banhara A, Ding Y, Kühner R, Zuccaro A, Parniske M . (2015). Colonization of root cells and plant growth promotion by Piriformospora indica occurs independently of plant common symbiosis genes. Front Plant Sci 6: 667.
Basiewicz M, Weiß M, Kogel K-H, Langen G, Zorn H, Zuccaro A . (2012). Molecular and phenotypic characterization of Sebacina vermifera strains associated with orchids, and the description of Piriformospora williamsii sp. nov. Fungal Biol 116: 204–213.
Behie SW, Zelisko PM, Bidochka MJ . (2012). Endophytic insect-parasitic fungi translocate nitrogen directly from insects to plants. Science 336: 1576–1577.
Bever JD, Dickie IA, Facelli E, Facelli JM, Klironomos J, Moora M et al. (2010). Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol 25: 468–478.
Bever JD, Platt TG, Morton ER . (2012). Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu Rev Microbiol 66: 265–283.
Blomberg SP, Garland T, Ives AR . (2003). Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57: 717–745.
Caldwell BA, Jumpponen A, Trappe JM . (2000). Utilization of major substrates by dark-septate, root endophytes. Mycologia 92: 230–232.
Carbonero F, Oakley BB, Purdy KJ . (2014). Metabolic flexibility as a major predictor of spatial distribution in microbial communities. PLoS One 9: e85105.
Castresana J . (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552.
Chagnon P-L, Bradley RL, Maherali H, Klironomos JN . (2013). A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant Sci 18: 484–491.
Cheikh-Ali Z, Glynou K, Ali T, Ploch S, Kaiser M, Thines M et al. (2015). Diversity of exophillic acid derivatives in strains of an endophytic Exophiala sp. Phytochemistry 118: 83–93.
Cohen J . (1988) Statistical Power Analysis for the Behavioral Sciences. L. Erlbaum Associates: Hillsdale, NJ.
Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T et al. (2016). Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209: 798–811.
Desdevises Y, Legendre P, Azouzi L, Morand S . (2003). Quantifying phylogenetically structured environmental variation. Evolution 57: 2647–2652.
Dufresne M, Osbourn AE . (2001). Definition of tissue-specific and general requirements for plant infection in a phytopathogenic fungus. Mol Plant Microbe Interact 14: 300–307.
Gardes M, Bruns TD . (1993). ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118.
Glynou K, Ali T, Buch A-K, Haghi Kia S, Ploch S, Xia X et al. (2016). The local environment determines the assembly of root endophytic fungi at a continental scale. Environ Microbiol 18: 2418–2434.
Hallmann J, Berg G, Schulz B . (2006) Isolation procedures for endophytic microorganisms. In: Schulz BJE, Boyle CJC, Sieber TN (eds). Microbial Root Endophytes, Soil Biology. Springer Berlin: Heidelberg, Germany, 299–319.
Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B et al. (2016). Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165: 464–474.
Hoeksema JD, Thompson JN . (2007). Geographic structure in a widespread plant–mycorrhizal interaction: pines and false truffles. J Evol Biol 20: 1148–1163.
Huelsenbeck JP, Ronquist F . (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755.
Ives AR, Midford PE, Garland T . (2007). Within-species variation and measurement error in phylogenetic comparative methods. Sys Biol 56: 252–270.
Johnsen HR, Krause K . (2014). Cellulase activity screening using pure carboxymethylcellulose: application to soluble cellulolytic samples and to plant tissue prints. Int J Mol Sci 15: 830–838.
Jumpponen A, Trappe JM . (1998). Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol 140: 295–310.
Katoh K, Standley DM . (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780.
Keim J, Mishra B, Sharma R, Ploch S, Thines M . (2014). Root-associated fungi of Arabidopsis thaliana and Microthlaspi perfoliatum. Fungal Divers 66: 99–111.
Klironomos JN . (2003). Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84: 2292–2301.
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22: 5271–5277.
Knapp DG, Pintye A, Kovács GM . (2012). The dark side is not fastidious—dark septate endophytic fungi of native and invasive plants of semiarid sandy areas. PLoS One 7: e32570.
Lahrmann U, Strehmel N, Langen G, Frerigmann H, Leson L, Ding Y et al. (2015). Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytol 207: 841–857.
Legendre P, Legendre LFJ . (2012) Numerical Ecology, Vol 24. 3rd edn, Elsevier: Amsterdam, Netherlands.
Maciá-Vicente JG, Ferraro V, Burruano S, Lopez-Llorca LV . (2012). Fungal assemblages associated with roots of halophytic and non-halophytic plant species vary differentially along a salinity gradient. Microb Ecol 64: 668–679.
Maciá-Vicente JG, Jansson H-B, Abdullah SK, Descals E, Salinas J, Lopez-Llorca LV . (2008a). Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiol Ecol 64: 90–105.
Maciá-Vicente JG, Jansson H-B, Mendgen K, Lopez-Llorca LV . (2008b). Colonization of barley roots by endophytic fungi and their reduction of take-all caused by Gaeumannomyces graminis var. tritici. Can J Microbiol 54: 600–609.
Maciá-Vicente JG, Jansson H-B, Talbot NJ, Lopez-Llorca LV . (2009a). Real-time PCR quantification and live-cell imaging of endophytic colonization of barley (Hordeum vulgare roots by Fusarium equiseti and Pochonia chlamydosporia. New Phytol 182: 213–228.
Maciá-Vicente JG, Rosso LC, Ciancio A, Jansson H-B, Lopez-Llorca LV . (2009b). Colonisation of barley roots by endophytic Fusarium equiseti and Pochonia chlamydosporia: effects on plant growth and disease. Ann Appl Biol 155: 391–401.
Maherali H, Klironomos JN . (2007). Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316: 1746–1748.
Maherali H, Klironomos JN . (2012). Phylogenetic and trait-based assembly of arbuscular mycorrhizal fungal communities. PLoS One 7: e36695.
Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, Di Pietro A et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373.
Mandyam K, Jumpponen A . (2005). Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud Mycol 53: 173–189.
Mandyam K, Jumpponen A . (2014) Unraveling the dark septate endophyte functions: insights from the Arabidopsis model. In: Verma VC, Gange AC (eds), Advances in Endophytic Research. Springer: India, 115–141.
Mandyam K, Jumpponen A . (2015). Mutualism–parasitism paradigm synthesized from results of root-endophyte models. Front Microbiol 5: 776.
Mandyam K, Loughin T, Jumpponen A . (2010). Isolation and morphological and metabolic characterization of common endophytes in annually burned tallgrass prairie. Mycologia 102: 813–821.
Mangan SA, Schnitzer SA, Herre EA, Mack KML, Valencia MC, Sanchez EI et al. (2010). Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466: 752–755.
Mayerhofer MS, Kernaghan G, Harper KA . (2012). The effects of fungal root endophytes on plant growth: a meta-analysis. Mycorrhiza 23: 119–128.
Moles AT, Ackerly DD, Webb CO, Tweddle JC, Dickie JB, Westoby M . (2005). A brief history of seed size. Science 307: 576–580.
Murashige T, Skoog F . (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497.
Newsham KK . (2011). A meta-analysis of plant responses to dark septate root endophytes. New Phytol 190: 783–793.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB et al. (2015). vegan: Community Ecology Package. Available from http://CRAN.R-project.org/package=vegan.
Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N . (2011) caper: Comparative Analyses of Phylogenetics and Evolution in R. Available from http://CRAN.R-project.org/package=caper.
Pagel M . (1999). Inferring the historical patterns of biological evolution. Nature 401: 877–884.
Peterson RL, Wagg C, Pautler M . (2008). Associations between microfungal endophytes and roots: do structural features indicate function? Botany 86: 445–456.
Piculell BJ, Hoeksema JD, Thompson JN . (2008). Interactions of biotic and abiotic environmental factors in an ectomycorrhizal symbiosis, and the potential for selection mosaics. BMC Biol 6: 23.
Powell JR, Parrent JL, Hart MM, Klironomos JN, Rillig MC, Maherali H . (2009). Phylogenetic trait conservatism and the evolution of functional trade-offs in arbuscular mycorrhizal fungi. Proc R Soc Lond B Biol Sci 276: 4237–4245.
R Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Redecker D, Hijri M, Dulieu H, Sanders IR . (1999). Phylogenetic analysis of a dataset of fungal 5.8 S rDNA sequences shows that highly divergent copies of internal transcribed spacers reported from Scutellospora castanea are of ascomycete origin. Fungal Genet Biol 28: 238–244.
Revell LJ . (2011). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3: 217–223.
Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2: 404–416.
Rodriguez RJ, White JF Jr, Arnold AE, Redman RS . (2009). Fungal endophytes: diversity and functional roles. New Phytol 182: 314–330.
Sacristán S, García-Arenal F . (2008). The evolution of virulence and pathogenicity in plant pathogen populations. Mol Plant Pathol 9: 369–384.
Sánchez Márquez S, Bills G, Herrero N, Zabalgogeazcoa I . (2012). Non-systemic fungal endophytes of grasses. Fungal Ecol 5: 289–297.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Sieber TN, Grünig CR . (2013) Fungal root endophytes. In: Plant Roots—The Hidden Half. CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 38-1–38-49.
Sperschneider J, Gardiner DM, Thatcher LF, Lyons R, Singh KB, Manners JM et al. (2015). Genome-wide analysis in three Fusarium pathogens identifies rapidly evolving chromosomes and genes associated with pathogenicity. Genome Biol Evol 7: 1613–1627.
Symonds MRE, Blomberg SP . (2014) A primer on phylogenetic generalised least squares. In : Garamszegi LZ (ed), Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology. Springer Berlin: Heidelberg, Germany, 105–130.
Tellenbach C, Grünig CR, Sieber TN . (2011). Negative effects on survival and performance of Norway spruce seedlings colonized by dark septate root endophytes are primarily isolate-dependent. Environ Microbiol 13: 2508–2517.
Thompson JN . (2005) The Geographic Mosaic of Coevolution. University of Chicago Press: Chicago, IL.
Torchiano M . (2015) effsize: Efficient Effect Size Computation. Available from http://CRAN.R-project.org/package=effsize.
Treseder KK, Lennon JT . (2015). Fungal traits that drive ecosystem dynamics on land. Microbiology and Mol Biol Rev 79: 243–262.
Usuki F, Narisawa K . (2007). A mutualistic symbiosis between a dark septate endophytic fungus, Heteroconium chaetospira, and a nonmycorrhizal plant, Chinese cabbage. Mycologia 99: 175–184.
Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JPW . (2002). Extensive fungal diversity in plant roots. Science 295: 2051–2051.
Van der Putten WH, Van Dijk C, Peters BAM . (1993). Plant-specific soil-borne diseases contribute to succession in foredune vegetation. Nature 362: 53–56.
Violle C, Jiang L . (2009). Towards a trait-based quantification of species niche. J Plant Ecol 2: 87–93.
Violle C, Navas M-L, Vile D, Kazakou E, Fortunel C, Hummel I et al. (2007). Let the concept of trait be functional!. Oikos 116: 882–892.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten WH, Wall DH . (2004). Ecological linkages between aboveground and belowground biota. Science 304: 1629–1633.
Webb CO, Ackerly DD, Kembel SW . (2008). Phylocom: software for the analysis of community phylogenetic structure and trait evolution. Bioinformatics 24: 2098–2100.
Wehner J, Powell JR, Muller LAH, Caruso T, Veresoglou SD, Hempel S et al. (2014). Determinants of root-associated fungal communities within Asteraceae in a semi-arid grassland. J Ecol 102: 425–436.
White TJ, Bruns T, Lee S, Taylor J . (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, Tj White (eds), PCR Protocols: a Guide to Methods and Applications. Academic Press: San Diego, CA, USA, pp 315–322.
Zavala-Gonzalez EA, Escudero N, Lopez-Moya F, Aranda-Martinez A, Exposito A, Ricaño-Rodríguez J et al. (2015). Some isolates of the nematophagous fungus Pochonia chlamydosporia promote root growth and reduce flowering time of tomato. Ann Appl Biol 166: 472–483.
Acknowledgements
We thank Johanna A Bac-Molenaar, Xiaojuan Xia and Saatzucht Josef Breun GmbH & Co. KG for kindly providing seeds of A. thaliana, M. erraticum and barley cv. Barke, respectively. We also thank Gábor M Kovács and three anonymous referees for their constructive comments on this manuscript. This study was supported by LOEWE (Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz) of the state of Hesse and was conducted within the framework of the Cluster for Integrative Fungal Research (IPF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Kia, S., Glynou, K., Nau, T. et al. Influence of phylogenetic conservatism and trait convergence on the interactions between fungal root endophytes and plants. ISME J 11, 777–790 (2017). https://doi.org/10.1038/ismej.2016.140
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.140
This article is cited by
-
Contrasting patterns in phylogenetic and biogeographic factories of invasive grasses (Poaceae) across the globe
npj Biodiversity (2023)
-
A fungal sesquiterpene biosynthesis gene cluster critical for mutualist-pathogen transition in Colletotrichum tofieldiae
Nature Communications (2023)
-
Genetic determinants of endophytism in the Arabidopsis root mycobiome
Nature Communications (2021)
-
Brassicaceous roots as an unexpected diversity hot-spot of helotialean endophytes
IMA Fungus (2020)
-
VBA–AMF: A VBA Program Based on the Magnified Intersections Method for Quantitative Recording of Root Colonization by Arbuscular Mycorrhizal Fungi
Indian Journal of Microbiology (2020)