Abstract
To probe the metabolic potential of abundant Archaea in boreal peats, we reconstructed two near-complete archaeal genomes, affiliated with Thaumarchaeota group 1.1c (bin Fn1, 8% abundance), which was a genomically unrepresented group, and Thermoplasmata (bin Bg1, 26% abundance), from metagenomic data acquired from deep anoxic peat layers. Each of the near-complete genomes encodes the potential to degrade long-chain fatty acids (LCFA) via β-oxidation. Fn1 has the potential to oxidize LCFA either by syntrophic interaction with methanogens or by coupling oxidation with anaerobic respiration using fumarate as a terminal electron acceptor (TEA). Fn1 is the first Thaumarchaeota genome without an identifiable carbon fixation pathway, indicating that this mesophilic phylum encompasses more diverse metabolisms than previously thought. Furthermore, we report genetic evidence suggestive of sulfite and/or organosulfonate reduction by Thermoplasmata Bg1. In deep peat, inorganic TEAs are often depleted to extremely low levels, yet the anaerobic respiration predicted for two abundant archaeal members suggests organic electron acceptors such as fumarate and organosulfonate (enriched in humic substances) may be important for respiration and C mineralization in peatlands.
Similar content being viewed by others
Main
Peatlands represent carbon rich environments shown to sequester approximately one-third of all soil carbon on Earth (Gorham, 1991). Archaea are particularly abundant in peat soils, with Crenarchaea and Thaumarchaeota comprising up to 60% of the microbial community in subsurface peats (Kemnitz et al., 2007; Lin et al., 2012; Basiliko et al., 2013; Hawkins et al., 2014; Lin et al., 2014). Thaumarchaeotal groups 1.1b and 1.1c were recently estimated to account for 76±33% of total archaea in global soil samples (Auguet et al., 2010), suggesting they may have key roles in Earth’s biogeochemical cycles. Currently, all cultured members of the Thaumarchaeota are autotrophic ammonia oxidizing archaea, belonging to group 1.1a and 1.1b (Konneke et al., 2005; de la Torre et al., 2008; Hatzenpichler et al., 2008; Tourna et al., 2011; Stieglmeier et al., 2014). The carbon metabolism and energy sources used by group 1.1c remain unknown. Characterization of uncultivated groups such as group 1.1c is essential to better understand the metabolic and evolutionary complexity of this phylum. Thermoplasmata represent another important component of soil microbial communities, accounting for ~5% of total archaeal sequences in global soil samples (Auguet et al., 2010). Poor genomic representation of both these archaeal groups makes it extremely difficult to assign unassembled whole-genome shotgun and transcriptomic sequences to these groups.
This study elucidates ecophysiological potential for archaeal taxa prevalent in peatlands using de novo assembly of metagenomic sequences from two deep peat column sections (Fen-75 cm and T3M-75 cm) that were collected at 75–100 cm depth from the Bog Lake Fen (47°30'22.62'', −93°29'20.46'') and S1 bog T3M (47°30'22.3914'', −93°27'11.772'') sites within the Marcell Experimental Forest, MN, USA. Extraction of genomic DNA, sequencing and bioinformatics analysis is described in detail in the Supplementary methods. Assembly resulted in the first reconstruction of two near-complete genomes for members of the Thaumarchaeota group 1.1c and Thermoplasmata terrestrial miscellaneous group (TMEG).
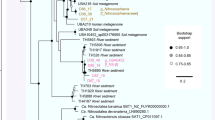
The Fen-75-cm and T3M-75-cm metagenomes contained six and four distinct genomic bins, respectively (Supplementary Table S1 and Supplementary Figures S1 and S2). Bins Fn1 and Bg1 representing two archaeal genomes are further characterized here, owing to their high abundance (Fn1, 8%; Bg1, 26% in relative abundance; Figure 1), genomic novelty and near completeness (Supplementary Table S1). Out of 104 archaeal marker genes, Fn1 had 95 and Bg1 had 102 (Supplementary Table S1), suggesting that each assembly likely represents a substantial fraction of a single draft genome. Phylogenetic analysis placed the Fn1 genome in the Thaumarchaeota group 1.1c and the Bg1 in the Thermoplasmata (Figure 1 and Supplementary Figure S3). Thus far, there are no cultured representatives or assembled genomes in group 1.1c and in Thermoplasmata TMEG. Therefore, our Fn1 and Bg1 genomes provide novel insights into the metabolic potential of these archaeal groups.
Maximum likelihood tree based on SSU rRNA gene sequences. Solid triangles, sequences retrieved from assembled contigs; open triangles, sequences reconstructed by EMIRGE (Expectation maximization iterative reconstruction of genes from the environment, see Supplementary methods). Numbers in parentheses indicate the percentage of sequences or read coverage.
A metabolic reconstruction for Fn1 revealed a set of typical anaerobic pathways (Figure 2 and Supplementary Table S2). A long-chain fatty acid (LCFA) β-oxidation cycle was predicted. An incomplete glycolysis pathway (Supplementary Figure S4), and the presence of genes encoding fructose-bisphosphatase and phosphoenolpyruvate carboxykinase suggested that Fn1 uses a gluconeogenesis pathway for synthesis of sugars (Pilkis and Claus, 1991). Fn1 did not contain genes encoding ATP citrate synthase and lyase (which catalyzes the cleavage of citrate into acetyl coenzyme A (acetyl-CoA) and oxaloacetate in a CoA- and ATP-dependent matter), suggesting that the TCA and reverse TCA cycles are not operative in Fn1. Instead, Fn1 contains an incomplete reverse TCA cycle as seen in methanogens (Goodchild et al., 2004). All six carbon fixation pathways known to date (Berg et al., 2010) were incomplete or undetectable in Fn1 (Supplementary Figure S5a). This latter feature is distinct from other sequenced thaumarchaeal and autotrophic crenarchaeal genomes (Berg et al., 2010; Beam et al., 2014; Konneke et al., 2014), further building on previous evidence which suggests a more versatile metabolism for members of the Phylum Thaumarchaeota (for example, bacterial ectosymbionts (Muller et al., 2010) and facultative ammonia oxidizers (Mussmann et al., 2011)). Fn1 is not predicted to ferment pyruvate to end products such as acetate, butyrate, ethanol, H2, lactate, and propionate (Supplementary Table S3). Regeneration of NAD+ and energy production in Fn1 likely relies on the β-oxidation cycle and an anaerobic electron transport chain (ETC) (Figure 2). The lack of a cytochrome bd/c-type terminal oxidase suggests that Fn1 does not respire oxygen. The presence of succinate dehydrogenase/fumarate reductase suggests that Fn1 may use fumarate as a TEA under anoxic conditions (Kröger, 1978; Maklashina et al., 1998), although the function of these genes (whether for fumarate reduction or succinate oxidation) cannot be deduced solely from the presented gene sequences and phylogenetic reconstruction (Supplementary Figure S6). All genes related to dissimilatory sulfate reduction and denitrification were absent, as were genes encoding glycoside hydrolase (GH) families for carbohydrate degradation (Supplementary Table S4). Ultimately, genomic inference suggests that fatty acids could be the major carbon and energy sources for Fn1. On the other hand, it should be stressed that predictions of gene function in Archaea are difficult and we cannot exclude the possibility that some metabolic functions might be encoded on extrachromosomal elements or were not detected as the genomes are not closed. Thus, experimental verification is needed to confirm the absence of functions.
Metabolic reconstruction of central carbon metabolism, electron transport chains, energy conservation pathways and ABC transporters deduced from Thaumarchaeota Fn1 and Thermoplasmata Bg1 genomes. Shared functions and intermediates are shown in black color. Blue and red colors highlighted those only found in Fn1 and Bg1, respectively. Dotted lines indicate possible electron flow for the Fn1 and Bg1 populations. Dashed lines indicate putative reactions or proton movement across the membrane. Gray arrows indicate missing functions. Numbers correspond to enzymes: 1, phosphoglucomutase; 2, glucokinase; 3, glucose-6-phosphate isomerase; 4, fructose-1,6-bisphosphatase; 5, 6-phosphofructokinase; 6, fructose-bisphosphate aldolase, class I; 7, glyceraldehyde-3-phosphate dehydrogenase (NAD(P)); 8, phosphoglycerate kinase; 9, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase; 10, enolase; 11, pyruvate kinase; 12, pyruvate ferredoxin oxidoreductase; 13, AMP-type acetyl-CoA synthetase; 14, phosphoenolpyruvate carboxykinase; 15, pyruvate carboxylase; 16, citrate synthase; 17, aconitate hydratase; 18, isocitrate dehydrogenase (NAD+); 19, 2-oxoglutarate ferredoxin oxidoreductase subunit alpha; 20, succinyl-CoA synthetase alpha subunit; 21, succinate dehydrogenase (ubiquinone) flavoprotein subunit; 22, fumarate hydratase; 23, malate dehydrogenase; 24, acetyl-CoA C-acetyltransferase; 25, aldehyde dehydrogenase (NAD+); 26, alcohol dehydrogenase; 27, transketolase; 28, ribulose-phosphate 3-epimerase; 29, ribose 5-phosphate isomerase; 30, ribose-phosphate pyrophosphokinase; 31, long-chain acyl-CoA synthetase; 32, acyl-coA dehydrogenase; 33, Enoyl-CoA hydratase; 34, 3-hydroxyacyl-CoA dehydrogenase; 35, 3-ketoacyl-CoA thiolase; 36, NADH-quinone oxidoreductase; 37, Succinate dehydrogenase/fumarate reductase; 38, ubiquinol-cytochrome c reductase; 39, F-type H+-transporting ATPase; 40, inorganic pyrophosphatase; 41, sulfate adenylyltransferase; 42, thiosulfate sulfurtransferase; 43, sulfur:oxygen oxidoreductase; 44, dissimilatory sulfite reductase. EC# for each enzyme is listed in Supplementary Table S2. Information on gene annotation, length, and annotation sources are listed in the Supplementary files ‘Supplementary information of genes annotated for the Fn1 genome.pdf’ and ‘Supplementary information of genes annotated for the Bg1 genome.pdf’.
Metabolic predictions from the Bg1 genome suggest a versatile heterotrophic lifestyle. The genome contains a complete glycolysis pathway, TCA cycle, β-oxidation cycle (Figure 2) and two types of GH families (GH15 and GH57; Supplementary Table S4), which respectively contain glucoamylases and alpha-amylase activities, as seen in the euryarchaeota, Pyrococcus furiosus (Laderman et al., 1993). Like Fn1, the Bg1 genome lacks key genes in any of the six carbon fixation pathways (Supplementary Figure S5b), suggesting obligate heterotrophy. Energy conservation likely relies on an anaerobic ETC, owing to the presence of 4 ETC complexes (I, II, III and V) (Figure 2). Genes encoding for dissimilatory adenylyl-sulfate reductase, thiosulfate reductase (Phs) and sulfur reductase (Sre) were absent, indicating that Bg1 is unable to respire these inorganic sulfur species. However, detection of genes encoding for a desulfoviridin (dissimilatory sulfite reductase, dsrAB) implies Bg1 uses sulfite as a TEA, generating sulfide in a 6-electron transfer. The presence of an alkanesulfonate transporter, but no identifiable alkanesulfonate monooxygenase, suggests Bg1 may have an unknown mechanism for degrading organosulfonate to sulfite, possibly in a similar manner to sulfonate, but non-sulfate, reducing Bilophila wadsworthia, (Laue et al., 2001). The predicted Bg1 dsrA protein sequence was most closely related to those of thermophilic Crenarchaeota such as Thermoproteus and Volcanisaeta, by <60% sequence identity (Supplementary Figure S7), and thus represents a phylogenetically novel wetland dsrA gene (Pester et al., 2012).
We predict LCFA degradation via a β-oxidation mechanism for both Thaumarcheaota Fn1 and Thermoplasmata Bg1. LCFA oxidation is thermodynamically impossible without coupling to respiration activity or the consumption of reducing equivalents (hydrogen/formate) (Schink, 1997). Considering the lack of genes related to sulfate/nitrate reduction in Fn1 and occurrence of abundant hydrogenotrophic methanogens in the deep peat (Lin et al., 2014), it is possible that Fn1 establishes a syntrophic interaction with methanogens and/or uses fumarate as a TEA to degrade LCFA. In addition, we reason that Thermoplasmata Bg1 may couple sulfite (and potentially sulfonate) reduction to LCFA oxidation, and organosulfonate is abundant in peat soils and humic substances (Autry and Fitzgerald, 1990) relative to the low levels of inorganic sulfate observed in peats.
References
Auguet JC, Barberan A, Casamayor EO . (2010). Global ecological patterns in uncultured Archaea. Isme J 4: 182–190.
Autry AR, Fitzgerald JW . (1990). Sulfonate-S - a Major Form of Forest Soil Organic Sulfur. Biol Fert Soils 10: 50–56.
Basiliko N, Henry K, Gupta V, Moore T, Driscoll B, Dunfield PF . (2013). Controls on bacterial and archaeal community structure and greenhouse gas production in natural,mined, and restored Canadian peatlands. Front Microbiol 4: 215.
Beam JP, Jay ZJ, Kozubal MA, Inskeep WP . (2014). Niche specialization of novel Thaumarchaeota to oxic and hypoxic acidic geothermal springs of Yellowstone National Park. ISME J 8: 938–951.
Berg IA, Ramos-Vera WH, Petri A, Huber H, Fuchs G . (2010). Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology+ 156: 256–269.
de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA . (2008). Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10: 810–818.
Goodchild A, Raftery M, Saunders NF, Guilhaus M, Cavicchioli R . (2004). Biology of the cold adapted archaeon, Methanococcoides burtonii determined by proteomics using liquid chromatography-tandem mass spectrometry. J Proteome Res 3: 1164–1176.
Gorham E . (1991). Northern peatlands: role in the carbon-cycle and probable responses to climatic warming. Ecol Appl 1: 182–195.
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H et al. (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA 105: 2134–2139.
Hawkins AN, Johnson KW, Brauer SL . (2014). Southern Appalachian peatlands support high archaeal diversity. Microb Ecol 67: 587–602.
Kemnitz D, Kolb S, Conrad R . (2007). High abundance of Crenarchaeota in a temperate acidic forest soil. Fems Microbiol Ecol 60: 442–448.
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Konneke M, Schubert DM, Brown PC, Hugler M, Standfest S, Schwander T et al. (2014). Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA 111: 8239–8244.
Kröger A . (1978). Fumarate as terminal acceptor of phosphorylative electron transport. Biochim Biophys Acta 505: 129–145.
Laderman KA, Davis BR, Krutzsch HC, Lewis MS, Griko YV, Privalov PL et al. (1993). The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Biol Chem 268: 24394–24401.
Laue H, Friedrich M, Ruff J, Cook AM . (2001). Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J Bacteriol 183: 1727–1733.
Lin X, Green S, Tfaily MM, Prakash O, Konstantinidis KT, Corbett JE et al. (2012). Microbial community structure and activity linked to contrasting biogeochemical gradients in bog and fen environments of the glacial lake agassiz peatland. Appl Environ Microbiol 78: 7023–7031.
Lin X, Tfaily MM, Steinweg JM, Chanton P, Esson K, Yang ZK et al. (2014). Microbial community stratification linked to utilization of carbohydrates and phosphorus limitation in a boreal peatland at Marcell Experimental Forest, Minnesota, USA. Appl Environ Microbiol 80: 3518–3530.
Maklashina E, Berthold DA, Cecchini G . (1998). Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J Bacteriol 180: 5989–5996.
Muller F, Brissac T, Le Bris N, Felbeck H, Gros O . (2010). First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol 12: 2371–2383.
Mussmann M, Brito I, Pitcher A, Sinninghe Damste JS, Hatzenpichler R, Richter A et al. (2011). Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA 108: 16771–16776.
Pester M, Knorr K-H, Friedrich M, Wagner M, Loy A . (2012). Sulfate-reducing microorganisms in wetlands – fameless actors in carbon cycling and climate change. Front Microbiol 3: 72.
Pilkis SJ, Claus TH . (1991). Hepatic gluconeogenesis/glycolysis: regulation and structure/function relationships of substrate cycle enzymes. Annu Rev Nutr 11: 465–515.
Schink B . (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61: 262–280.
Stieglmeier M, Klingl A, Alves RJ, Rittmann SK, Melcher M, Leisch N et al. (2014). Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol 64: 2738–2752.
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108: 8420–8425.
Acknowledgements
We thank Randall K. Kolka, USDA Forest Service, Paul J. Hanson, Colleen Iversen and other members of the SPRUCE project team for facilitating access to the Marcell Experimental Forest and the SPRUCE experimental facilities, and for helping with sample handling and laboratory space support. We thank P. Chanton and K. Esson for peat soil sampling, and S. Owens for the Illumina library preparation and sequencing. We acknowledge the University of Chicago Research Computing Center for support of this work. This work was supported by the Office of Biological and Environmental Research, Terrestrial Ecosystem Science Program, under U.S. DOE contracts DE-SC0007144 and DE-SC0012088, the U.S. National Science Foundation (NSF-EAR-0628349), and in part by the U.S. DOE contract DE-AC02-06CH11357. We wish to thank the generous support of the Alfred P Sloan Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Lin, X., Handley, K., Gilbert, J. et al. Metabolic potential of fatty acid oxidation and anaerobic respiration by abundant members of Thaumarchaeota and Thermoplasmata in deep anoxic peat. ISME J 9, 2740–2744 (2015). https://doi.org/10.1038/ismej.2015.77
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.77
This article is cited by
-
Metagenomic characterization of a novel non-ammonia-oxidizing Thaumarchaeota from hadal sediment
Microbiome (2024)
-
Anaerobic degradation of organic carbon supports uncultured microbial populations in estuarine sediments
Microbiome (2023)
-
Diverse ecophysiological adaptations of subsurface Thaumarchaeota in floodplain sediments revealed through genome-resolved metagenomics
The ISME Journal (2022)
-
Community structure and activity potentials of archaeal communities in hadal sediments of the Mariana and Mussau trenches
Marine Life Science & Technology (2022)
-
Biohythane production from tofu processing residue via two-stage anaerobic digestion: operational conditions and microbial community dynamics
Biomass Conversion and Biorefinery (2022)