Abstract
The genus Bradyrhizobium has served as a model system for studying host–microbe symbiotic interactions and nitrogen fixation due to its importance in agricultural productivity and global nitrogen cycling. In this study, we identify a bacterial group affiliated with this genus that dominates the microbial communities of coniferous forest soils from six distinct ecozones across North America. Representative isolates from this group were obtained and characterized. Using quantitative population genomics, we show that forest soil populations of Bradyrhizobium represent ecotypes incapable of nodulating legume root hairs or fixing atmospheric nitrogen. Instead, these populations appear to be free living and have a greater potential for metabolizing aromatic carbon sources than their close symbiotic relatives. In addition, we identify fine-scaled differentiation between populations inhabiting neighboring soil layers that illustrate how diversity within Bradyrhizobium is structured by habitat similarity. These findings reconcile incongruent observations about this widely studied and important group of bacteria and highlight the value of ecological context to interpretations of microbial diversity and taxonomy. These results further suggest that the influence of this genus likely extends well beyond facilitating agriculture, especially as forest ecosystems are large and integral components of the biosphere. In addition, this study demonstrates how focusing research on economically important microorganisms can bias our understanding of the natural world.
Similar content being viewed by others
Introduction
Members of Bradyrhizobium have been extensively characterized for their ability to fix nitrogen and enter the root hairs of leguminous plants to form symbiotic nodules. These nodules provide the host plant with a source of fixed nitrogen in exchange for a portion of its photosynthetic products. This mutualistic interaction serves important roles in global nitrogen cycling and modern agriculture (Zahran, 1999). For these reasons, members of this genus are studied extensively as a model of legume–rhizobia symbioses and widely acknowledged to be ecologically and economically important organisms.
However, culture-independent surveys of bacterial communities have identified Bradyrhizobium populations in soil habitats distinct from the rhizosphere of leguminous plants (Uroz et al., 2010; Delmont et al., 2012; Hartmann et al., 2012). Nevertheless, the hypothesized ecological roles of these populations remain focused on symbiosis. Similarly, previous studies have isolated Bradyrhizobium strains diminished or lacking in the ability to fix nitrogen or nodulate leguminous plants from soil, but these strains were described consistently as having transiently abandoned a symbiotic lifestyle or as relics of an ancestral symbiotic genotype (Sachs et al., 2010, 2011; Okubo et al., 2012).
It is widely recognized that the nitrogen fixation (nif) and nodulation (nod) gene clusters often occur within genomic islands or on extrachromosomal elements and show strong evidence of non-vertical descent among the Bradyrhizobia (MacLean et al., 2007). Yet, the inability to fix nitrogen or nodulate legumes has been interpreted as recent and transient divergence from the group, as opposed to examples of stable differentiation within it. Different patterns of environmental selection between habitats can cause considerable divergence in the ecological properties of closely related populations, even between populations with identical marker gene sequences (Shapiro and Polz, 2014). So, although this symbiotic lifestyle is attributed to soil Bradyrhizobia, it is possible that the strains and populations that do not fit this rigid definition are non-symbiotic ecotypes that occupy physically or functionally distinct niches.
Here we identify and characterize an operational taxonomic unit (OTU) affiliated with the genus Bradyrhizobium that dominates the microbial communities of coniferous forest soils across North America. Using quantitative population genomics, we reconcile the widespread abundance of this group in soils lacking leguminous plants with the long-standing association of this genus with legume symbiosis. By determining the abundance of the shared and non-shared genomic regions among four representative isolates from this OTU with respect to their closest symbiotic relatives, we show that forest soil populations are ecologically distinct from symbiotic populations. Furthermore, we demonstrate how diversity within Bradyrhizobium is structured by habitat similarity.
Materials and methods
Soil samples
Samples were collected from experimental managed forests at 18 sites, three in each of six ecozones across North America, representing distinct climatic regimes (Supplementary Table S1), which are part of the long-term soil productivity (LTSP) study (Powers, 2006). Approximately 51 samples were collected at each site. Soils were sampled as previously described (Hartmann et al., 2012). At each sample point, both the organic layer (forest floor) and top 20 cm of the mineral layer were collected separately. Each sample was a composite of three to five samples. The composites were mixed in the field, and a subsample was collected in either 50-ml conical tubes or plastic bags. Samples were stored at 4 °C during transport and until they could be processed. Soils were sieved through 2-mm mesh to remove roots and to further homogenize samples before subdividing each for analyses. The samples were stored at −80 °C.
DNA extraction
DNA was extracted from 0.5 g of sieved soil using the FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA) with bead matrix E according to the manufacturer’s protocol. DNA extracts were quantified using Quant-iT PicoGreen kit (Invitrogen, Carlsbad, CA, USA).
PCR amplification and sequencing of taxonomic markers
Partial bacterial 16 S rRNA genes (V1–V3) and fungal internal transcribed spacers (ITS2) were amplified using 10 ng of extracted DNA as template. The V1–V3 was amplified using 27F and 519R primers (Lane, 1991) with Roche 454-Titanium Flex plus adapters and MID barcodes for sequencing in the 5′ to 3′ direction. The ITS2 was amplified using ITS3 and ITS4 primers (White et al., 1990) also with sequencing adapters and barcodes for sequencing in the 3′ to 5′ direction. Amplification was in triplicate using the HotStar Taq amplification kit (Qiagen, Mississauga, ON, USA) with the addition of 15 μg bovine serum albumin. After 10 min at 95 °C to activate the polymerase, the reaction was run for 30 cycles of 95 °C for 30 s, 49 °C for 30 s and 72 °C for 90 s, followed by a final extension at 72 °C for 10 min. Negative PCR controls were run for each barcode. Triplicate reactions were pooled before purification using the Agencourt AMPure XP magnetic bead kit (Agencourt A63881, Brea, CA, USA). The manufacturer’s protocol was modified by cleaning four times as much PCR product as recommended (100 μl PCR product using 20 μl beads) and adding 50 μl of 20% polyethylene glycol 6000/0.9 m NaCl to accommodate the increased volume. The amount of extra PEG added was determined experimentally to maximize retention of DNA fragments greater than 200 bp, while preventing carryover of fragments 100 bp and smaller. Purified amplicons were quantified again with the PicoGreen kit and diluted to equimolar concentrations before submission to Genome Quebec for 454-Titanium pyrosequencing with 40 samples pooled on each half plate, which yielded ~500 000 total sequence reads.
Pyrotag analysis
Sequence processing was conducted using Mothur v.1.28 following the 454 SOP (Schloss et al., 2011) with some modifications. Barcode and primer matching and length screening allowed one mismatch for barcodes, two for primer and a minimum flow length of 300 for bacterial sequences. After denoising, all sequences shorter than 200 bp were discarded. Bacterial sequences were aligned against the dereplicated Silva rRNA database (v102) and trimmed to a common alignment position. Chimera checking was conducted using the uchime algorithm with the sequences as their own references and all putative chimeras were discarded. All sequences were classified using the Silva database as the taxonomic reference. Sequences were split by phylum identification before clustering, and sequences within each phylum were separately clustered to generate 97% identical OTUs. Taxonomic analyses were perfomed by classifying all unique sequences against the Silva database. Sequences are in the supplement of Hartmann et al. (2012) and in the Short read archive under study accession PRJEB8599, sample accessions ERS662612 to ERS663023.
Phylogenetic analyses
Whole-genome phylogeny analysis was performed as previously described (Avrani et al., 2011) using custom python scripts. The predicted protein sequences of each genome were compared with those of the other genomes using BLASTP (Altschul et al., 1997); reciprocal best blast hits of ⩾50% identity, aligned over ⩾70% of the length of both query and subject sequences were defined as orthologous genes. The distance between each pair of genomes was calculated as the number of shared orthologs divided by the number of protein-coding genes in the smaller genome. The distances were used to construct a neighbor-joining tree using the R package, phangorn, and bootstrap values of the tree topology were calculated using the R package, ape, based on 1000 resamplings. The maximum likelihood phylogeny of the LTSP isolates relative to all available publicly sequenced Bradyrhizobium strains was constructed using the DNA-directed RNA polymerase beta subunit protein sequence. RNA polymerase beta subunit was chosen for its greater ability to distinguish strains than 16S rRNA gene sequences. Sequences were aligned using MUSCLE (Edgar, 2004), and the ML tree was constructed using MEGA6.0 (Tamura et al., 2013).
Genome sequencing and analysis
The LTSP strain genomes were sequenced and assembled at the Michael Smith Genome Sciences Centre, Vancouver, Canada (LTSP strains 849 and M299) and at Genome Quebec, Montréal, Québec (LTSP strains 857 and 885). The genome sequences are available under bioproject PRJNA275239, sample accessions SAMN03340218, SAMN03340243, SAMN03340244 and SAMN03340245. The genomes were annotated using the integrated microbial genomes (IMGs) database and comparative analysis system (Markowitz et al., 2014). All reference genomes, including predicted ORFs and annotations, were downloaded from IMG. To identify shared and non-shared genome regions between genomes, the pairs of orthologous genes between genomes (defined during the construction of the whole-genome phylogeny analysis) were used to identify regions containing 10 or more consecutive non-orthologous genes. Pathway prediction was performed using Pathway Tools (Menlo Park, CA, USA) Software Version 17.0 using the IMG annotations (Dale et al., 2010). All reported pathways were manually curated to ensure that they were not erroneously called. CAZy class assignment was performed by comparing the predicted proteins with the database of CAZy proteins using BLASTP. All predicted proteins with hits to CAZymes (⩾50% identity and aligned over ⩾70% of the length of both query and subject sequences) were classified as members of the CAZy class of their top hit.
Metagenome sequencing and analysis
Shotgun metagenomes were sequenced at the Michael Smith Genome Sciences Centre, Vancouver, Canada. Two sets of triplicate samples from the O’Connor Lake LTSP site in the IDF ecozone were analyzed. Each set of triplicates was barcoded, pooled and sequenced on a single lane of an Illumina Hiseq 2000 (San Diego, CA, USA). Sequencing generated 75-base paired-end reads from library inserts ranging from 80 to 214 bp. Raw reads were filtered with the NGS QC toolkit v2.3 (Patel and Jain, 2012) using a cutoff PHRED quality value of 20 over 70% of the read length. The organic layer metagenome contained 171 738 664 sequences (8.7 Gbases), and the mineral layer metagenome contained 158 694 371 sequences (9.5 Gbases). These metagenomic reads are available at the Short read archive under study accession PRJEB8420, sample accessions ERS656890 to ERS656895. Short metagenomic reads were compared with the predicted genes of the genomes of reference strains and LTSP isolates using BLASTN (Altschul et al., 1997) with an E-value threshold of 1E−10. Genes differentially enriched in the mineral versus organic soil layers were identified, similarly to as previously described (Coleman and Chisholm, 2010b). The distance of each gene from equal abundance in both metagenomes was calculated using the following equation:

Where D is distance and A is abundance of the gene in each soil layer metagenome. A P-value for enrichment in the mineral layer metagenome was calculated by comparing the distances of non-shared genes with the distribution of distances observed among the core genes shared by both strains. These calculated P-values of enrichment were false-discovery rate corrected, and significantly enriched genes were defined as those with a false discovery rate q-value ⩽0.01.
Results and Discussion
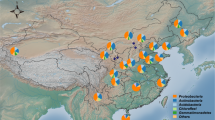
In association with the Long-Term Soil Productivity (LTSP) study, we surveyed the bacterial communities of forest soils located in six distinct ecozones across North America by sequencing the hypervariable region V1–V3 of the bacterial 16S gene and clustering OTUs at 97% identity (Figure 1a). Although each of the forests we studied had different successional histories and climatic conditions (Figure 1b; Supplementary Table S1), nearly every sample we collected was dominated by one OTU affiliated with the genus Bradyrhizobium (OTU1; Figure 1c). In fact, OTU1 often outnumbered the next most abundant OTU by an order of magnitude. It is unusual for an OTU to be so ubiquitous and at such high relative abundance in forest soils and soils in general (Schloss and Handelsman, 2006). Similar studies of coniferous forest soils have reported the most abundant OTU to occupy 2–8% of the community (Baldrian et al., 2012; Williams et al., 2013). However, results concerning OTU1 were independent of the methods used to process and cluster the sequences and the PCR primers used, and the relative abundance of this OTU was separately validated using quantitative PCR (Hartmann et al., 2012). Furthermore, the identity of OTU1 was surprising, as soil samples for this study were collected from bulk, rather than rhizosphere soil, and the sites had very few leguminous plants. The remarkable abundance and intriguing taxonomic affiliation of OTU1 prompted us to isolate representative strains to investigate their ecology (VanInsberghe et al., 2013). Specifically, we were interested in determining whether there are alternative lifestyles that populations within this group occupy besides those associated with the genus.
OTU1 is the most abundant group of bacteria in the pyrotag data set of nearly every sample collected from 18 forest sites in six ecozones across North America. (a) The locations of the three sites sampled in each ecozone, with ecozones named after their dominant tree species. (b) Ecozone climatic information and dominant tree species name. (c) The relative abundance of OTU1 in the pyrotag data set of each sample surveyed. OTU1 dominates the bacterial community in 95% of the 737 samples.
Only one previous study reported the isolation from soil of a Bradyrhizobium strain that lacks the common nod genes (strain S23321; Figure 2), and none have reported strains lacking the nif genes (Okubo et al., 2012). This suggested that OTU1 may be an important component of the nitrogen cycle in these soils. However, none of the genomes of the four LTSP OTU1 isolates that we examined possessed the nif or nod gene clusters, despite their close phylogenetic relationship with type strains from genus Bradyrhizobium (Supplementary Figure S1). In fact, by identifying regions of shared gene content with closely related symbiotic strains, we found that entire genomic regions, including those containing the nif and nod gene clusters, are absent from the LTSP isolates (Figures 2a–c; Supplementary Figures S2a–c). It is unlikely that we overlooked divergent orthologs of these regions, because our comparative method was able to detect the nif and nod gene clusters in distantly related Bradyrhizobium species.
Bradyrhizobium populations that dominate LTSP sites are not capable of nodulation or nitrogen fixation. (a) Genomic content of four strains compared with that of B. japonicum USDA 110; core regions are those shared by all five strains. (b) The percent identity of the reciprocal best blast hits (RBHs) in the four query genomes are plotted relative to their location in the USDA 110 genome. Gaps between RBHs indicate genomic islands present in the USDA 110 genome but absent from the respective query strain. (c) Percent GC in the USDA 110 genome indicates that the symbiosis island was likely acquired recently. (d) The moving average of the number of metagenomic reads from LTSP test plots mapped to each ORF in the USDA 110 genome by location is shown. This indicates that the LTSP strains are representative of the Bradyrhizobium spp. in the forest soils, as the genes in the symbiosis island are much lower in abundance than those in the shared backbone.
To test whether the four LTSP isolates are representative of their populations in situ, and not atypical variants that lack the nif and nod gene clusters, we mapped metagenomic reads from LTSP soils onto the genomes of reference strains most closely related to the isolates (Figure 2d; Supplementary Figure S2d). Just as there is an absence of orthologous genes in the genomes of the LTSP strains across the entire symbiosis island, these regions recruited about two orders of magnitude fewer metagenomic reads per gene than did the core genome. In fact, substantially fewer reads mapped to all of the genomic regions absent in the LTSP strains. This indicates that the Bradyrhizobium populations that dominate forest soils are not endosymbiotic nitrogen fixers, and instead represent unique ecotypes. Despite their abundance, we are unable to comment on the activity of the non-symbiotic populations relative to their symbiotic counterparts, as a large proportion of the former may be dormant.
The conclusion that OTU1 and the LTSP isolates represent free-living ecotypes distinct from the classical symbiotic ecotype is further supported by substantially different suites of metabolic genes in the two groups (Figure 3). The largest metabolic distinctions between ecotypes are a larger set of aromatic degradation pathways in the free-living strains and larger sets of nitrogen metabolism and polymer degradation genes in the symbiotic strains. Thus, the free-living ecotype may be adapted for using aromatic soil components. Notably, there is no identifiable relationship between genome size and the potential for symbiosis (Figure 3).
Genomic content of strains isolated from LTSP plots is substantially different from that of previously isolated relatives. (a) Phylogram shows relatedness between strains isolated from LTSP sites (in bold) and previously isolated strains based on whole-genome phylogeny; distance is proportional to the number of genes shared between strains, and bootstrap values are shown in red. Strain pairs LTSP 885 and LTSP M299 as well as LTSP 849 and LTSP 857 have identical 16S rRNA genes. (b) Occurrence of genes encoding metabolic pathways predicted using PathwayTools and curated manually and the relative abundance of carbohydrate active enzyme (CAZy) gene classes.
Looking closer, we see that the diversity within Bradyrhizobium goes beyond a simple division between symbiotic and free-living populations. There is a surprisingly large amount of genomic divergence between strains LTSP 885 and LTSP M299, considering their identical 16 S rRNA genes (Figure 3). These strains, respectively, were isolated from the organic and mineral soil layers at the same study site. By determining the abundance of each predicted gene in the genome of strain LTSP M299 in mineral and organic layer metagenomes, we found 133 genes significantly enriched in the mineral layer (Figure 4a). Interestingly, the majority of these genes (103 of 133) occur in genomic islands containing 10 or more genes that are not present in LTSP 885 (Figure 4b). The occurrence of these genes in discrete clusters suggests that the clusters were acquired as units and encode functions that are adaptive in the mineral layer. Annotations of the genes in these clusters provide further support for this hypothesis. For instance, a putative nitrogen scavenging cluster (Figure 4b and Supplementary Table S2) may be adaptive in the mineral layer, which has substantially lower mineralizable nitrogen concentrations than the organic layer (Supplementary Figure S3). Together, these results suggest that LTSP 885 and LTSP M299 represent neighboring, but distinct populations that have recently diverged, or are in the process of diverging due to differential selective pressures in the two soil layers.
Bradyrhizobium sp. LTSP M299 contains genes enriched in the mineral soil layer, typically occurring in clusters on genomic islands or putative extrachromosomal elements. (a) The abundance of LTSP M299 ORFs in metagenomes from mineral and organic soil layers shows that most of the ORFs enriched in the mineral layer (FDR-q value ⩽0.01) are not shared with the closely related strain LTSP 885 isolated from the organic layer. (b) Gaps between RBHs indicate genomic regions present in the LTSP M299 genome but absent from LTSP 885, and vertical red lines indicate ends of contigs. Most of the ORFs enriched in the mineral layer (103 of 133) occur in genomic regions containing 10 or more ORFs not shared with LTSP 885, often in discrete clusters within these regions. Percent GC, frequency of tetra-A homopolymer regions provide supporting evidence that the non-shared genomic regions were acquired by horizontal transfer.
By examining all currently available Bradyrhizobium genomes, we find that the absence of nif genes is actually widespread and not limited to the LTSP isolates (Supplementary Figure S1). Of 44 members of Bradyrhizobium whose genomes were sequenced before this study, seven lack a nif gene cluster, despite being isolated primarily from soils and root nodules. It is possible that some of these strains represent additional ecotypes with traits distinct from both the classical endosymbiotic and forest soil ecotypes. Because of the complexity of the soil environment, it is also possible that further novel ecotypes exist within the same soils where endosymbiotic populations are prevalent, occupying distinct physical or functional niche space.
The results reported here strongly support a growing body of literature that has demonstrated the utility of studying microbial diversity at the population level (Coleman and Chisholm, 2010a; Shapiro et al., 2012). A commonly overlooked subtlety of microbial speciation is that it will more often initiate than proceed to completion (Mallet, 2008). The barriers that promote divergence of populations by constraining gene flow in the absence of total genetic isolation, such as micro-geographic separation, are thought to be more important for structuring microbial diversity than those that lead to complete isolation. Thus, defining the boundaries of microbial species may not be as informative as identifying groups of co-existing individuals that share a common gene exchange network and key ecological features (Cordero and Polz, 2014; Shapiro and Polz, 2014).
The view that Bradyrhizobium is principally a lineage of nitrogen-fixing legume symbionts has likely been perpetuated in part by the practice of isolating strains from the rhizosphere and nodules of legumes using media that select for nitrogen-fixing heterotrophs. By isolating Bradyrhizobium strains from soils with few legumes, using dilute media with a source of fixed nitrogen (VanInsberghe et al., 2013), we accessed a previously unrecognized portion of the functional diversity in this taxonomic group present in soils. Had we attempted to isolate Bradyrhizobium strains using nitrogen-free media, it is likely that we would have isolated representatives of nitrogen-fixing populations that are at least 100-fold lower in abundance in these soils than the non-symbiotic populations.
Having explored differences between symbiotic and forest soil Bradyrhizobium populations and between forest soil populations from adjacent soil layers, we see how diversity in this genus is structured by habitat similarity. Given the prevalence and dominance of these forest soil ecotypes, our results suggest further that symbiosis may not be the dominant lifestyle within Bradyrhizobium, but rather one form of specialization within it. It is likely that more taxa currently regarded as well characterized will emerge as having unexpected amounts of ecological flexibility as their diversity is further explored in novel habitats. As the extent of this ecological flexibility becomes clear, our understanding of how microbes affect us will change as our knowledge becomes less biased by the study of economically important microbes. Embracing this feature of microbial diversity in future studies will ultimately lead to a more accurate, richer and more compelling vision of microbial systems and evolution.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D . (2011). Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474: 604–608.
Baldrian P, Kolařík M, Stursová M, Kopecký J, Valášková V, Větrovský CT et al. (2012). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6: 248–258.
Coleman ML, Chisholm SW . (2010a). Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA 107: 18634–18639.
Coleman ML, Chisholm SW . (2010b). Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA 107: 18634–18639.
Cordero OX, Polz MF . (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12: 263–273.
Dale JM, Popescu L, Karp PD . (2010). Machine learning methods for metabolic pathway prediction. BMC Bioinformatics 11: 15.
Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM et al. (2012). Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J 6: 1677–1687.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Hartmann M, Howes CG, VanInsberghe D, Yu H, Bachar D, Christen R et al. (2012). Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J 6: 2199–2218.
Lane DJ . (1991). 16 S/23 S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons: New York, pp 115–175.
MacLean AM, Finan TM, Sadowsky MJ . (2007). Genomes of the symbiotic nitrogen-fixing bacteria of legumes. Plant Physiol 144: 615–622.
Mallet J . (2008). Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos Trans R Soc Lond B Biol Sci 363: 2971–2986.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42: D560–D567.
Okubo T, Tsukui T, Maita H, Okamoto S, Oshima K, Fujisawa T et al. (2012). Complete Genome Sequence of Bradyrhizobium sp. S23321: Insights into Symbiosis Evolution in Soil Oligotrophs. Microbes Environ 27: 306–315.
Patel RK, Jain M . (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619.
Powers RF . (2006). Long-Term Soil Productivity: genesis of the concept and principles behind the program. Can J For Res 36: 519–528.
Sachs JL, Ehinger MO, Simms EL . (2010). Origins of cheating and loss of symbiosis in wild Bradyrhizobium. J Evol Biol 23: 1075–1089.
Sachs JL, Russell JE, Hollowell AC Evolutionary instability of symbiotic function in Bradyrhizobium japonicum. Dutilh BE (ed). PLoS One 2011; 6: e26370.
Schloss PD, Gevers D, Westcott SL Reducing the effects of PCR amplification and sequencing artifacts on 16 S rRNA-based studies. Gilbert JA (ed). PLoS One 2011; 6: e27310.
Schloss PD, Handelsman J . (2006). Toward a census of bacteria in soil. PLoS Comput Biol 2: e92.
Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G et al. (2012). Population genomics of early events in the ecological differentiation of bacteria. Science 336: 48–51.
Shapiro BJ, Polz MF . (2014). Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol 22: 235–247.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729.
Uroz S, Buée M, Murat C, Frey-Klett P, Martin F . (2010). Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2: 281–288.
VanInsberghe D, Hartmann M, Stewart GR, Mohn WW . (2013). Isolation of a substantial proportion of forest soil bacterial communities detected via pyrotag sequencing. Appl Environ Microbiol Microbiol 79: 2096–2098.
White T, Bruns T, Lee S, Taylor J . (1990). Amplification and direct sequencing of fungal ribosomal RNA for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T (eds). PCR Protocols: a Guide to Methods and Applications. Academic Press: New York, pp 315–322.
Williams MA, Jangid K, Shanmugam SG, Whitman WB . (2013). Bacterial communities in soil mimic patterns of vegetative succession and ecosystem climax but are resilient to change between seasons. Soil Biol Biochem 57: 749–757.
Zahran HH . (1999). Rhizobium-Legume Symbiosis and Nitrogen Fixation under Severe Conditions and in an Arid Climate. Microbiol Mol Biol Rev 63: 968–989.
Acknowledgements
This study was supported by grants from Genome Canada, Genome BC and the Tula Foundation. The Western Canadian Research Grid (WestGrid) provided access to high-performance computing resources. We thank Gordon R Stewart and Larisa O’Neal for the technical support as well as Martin F Polz and Sallie W Chisholm for useful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
VanInsberghe, D., Maas, K., Cardenas, E. et al. Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J 9, 2435–2441 (2015). https://doi.org/10.1038/ismej.2015.54
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.54
This article is cited by
-
Organic matter decomposition and associated microbial communities in wetlands: insights from tropical and subtropical Melaleuca forests in Australia
Hydrobiologia (2024)
-
The soil microbiomes of forest ecosystems in Kenya: their diversity and environmental drivers
Scientific Reports (2023)
-
Analysis of the speciation process suggests a dual lifestyle in the plant pathogen Ralstonia solanacearum species complex
European Journal of Plant Pathology (2023)
-
Multifarious Responses of Forest Soil Microbial Community Toward Climate Change
Microbial Ecology (2023)
-
Understory vegetation restoration improves soil physicochemical properties, enzymatic activity, and changes diazotrophic communities in Cunninghamia lanceolata plantations but depends on site history
Plant and Soil (2023)