Abstract
Mid-domain effect (MDE) models predict that the random placement of species’ ranges within a bounded geographical area leads to increased range overlap and species richness in the center of the bounded area. These models are frequently applied to study species-richness patterns of macroorganisms, but the MDE in relation to microorganisms is poorly understood. In this study, we examined the characteristics of the MDE in richness patterns of ectomycorrhizal (EM) fungi, an ecologically important group of soil symbionts. We conducted intensive soil sampling to investigate overlap among species ranges and the applicability of the MDE to EM fungi in four temperate forest stands along an elevation gradient on Mount Fuji, Japan. Molecular analyses using direct sequencing revealed 302 EM fungal species. Of 73 EM fungal species found in multiple stands, 72 inhabited a continuous range along the elevation gradient. The maximum overlap in species range and the highest species richness occurred at elevations in the middle of the gradient. The observed richness pattern also fit within the 95% confidence interval of the mid-domain null model, supporting the role of the MDE in EM fungal richness. Deviation in observed richness from the mean of the mid-domain null estimation was negatively correlated with some environmental factors, including precipitation and soil C/N, indicating that unexplained richness patterns could be driven by these environmental factors. Our results clearly support the existence of microbial species’ ranges along environmental gradients and the potential applicability of the MDE to better understand microbial diversity patterns.
Similar content being viewed by others
Introduction
Ectomycorrhizal (EM) fungi are plant–root symbionts that have critical roles in forest functioning, especially in nutrient cycling. These fungi establish obligate associations with many species of host trees and receive up to 22% of the photosynthetic carbon produced by these trees (Hobbie, 2006). In return, the extensive mycelial networks of EM fungi absorb soil nutrients and can deliver these nutrients, which may comprise >40% of plant nitrogen, to their hosts (Hobbie and Hobbie, 2006). The diversity and abundance of EM fungi are thought to influence tree growth and establishment, and thus overall forest health (Horton et al., 1999; Nara, 2006). EM fungi are taxonomically and functionally diverse and show high sensitivity to environmental change (Tedersoo et al., 2010; Rineau and Courty, 2011). Therefore, better understanding of these fungal communities is critical for conservation of biodiversity under environmental change and forest degradation (Courty et al., 2010; Koide et al., 2011).
The advancement of molecular techniques has improved accurate identification of microorganisms in field samples and has revealed the existence of highly diverse EM fungal communities in many ecosystems (Horton and Bruns, 2001; Ishida et al., 2007; Peay et al., 2010). Numerous studies have reported that EM fungal communities are structured by both biotic and abiotic factors, including host plants (Ishida et al., 2007; Tedersoo et al., 2008; Murata et al., 2013), soil environmental factors (e.g., pH, nutrients and litter characteristics) (Aponte et al., 2010; Cox et al., 2010; Peay et al., 2010), climate (Bahram et al., 2012), succession (Visser, 1995; Nara et al., 2003; Twieg et al., 2007) and competition (Pickles et al., 2012). However, little is known about geographic patterns in species richness of EM fungi or the mechanisms underlying these patterns.
Numerous studies have demonstrated geographical patterns of species richness in plants and animals. Species richness of most macroorganism groups peaks in tropical latitudes and decreases toward the poles (Gaston, 2000; Hillebrand, 2004), or peaks at intermediate elevations and decreases toward lower and higher elevations (Grytnes, 2003; McCain, 2004; Rahbek, 2005). Although the causal mechanisms for these general patterns remain controversial, mid-domain effect (MDE) models have been repeatedly proposed as a candidate mechanism to explain observed patterns or as a null model for extracting causal factors from the patterns (Colwell et al., 2004; Currie and Kerr, 2008). MDE models predict that random placement of species ranges within a bounded geographical area (i.e., a domain) leads to maximum overlap in species ranges and to the highest richness at mid-domain locations, excluding environmental and biological factors (Colwell and Lees, 2000).
Elevation gradients provide an ideal opportunity for testing the applicability of MDE models (Kluge et al., 2006; McCain, 2009; Lee et al., 2013). Species distributions and range overlaps across sites are readily recognized in plants and animals, but examining such patterns in soil microorganisms remains a major challenge. Inventories of EM fungi in microbial communities are often incomplete because these communities generally include many rare species that are likely to be overlooked in general surveys (Horton and Bruns, 2001; Peay et al., 2008). Thus, EM communities reported in the literature represent small fractions of the species existing at the studied sites. To our knowledge, no studies have quantified range overlap, and thus the applicability of the MDE, among EM fungal species along elevation gradients.
Although many different sampling methods have been used to study EM fungal communities, a clear tradeoff exists between the number of samples collected and the number of sites surveyed. Extensive sampling methods (i.e., more sampling sites but fewer samples per site) tend to result in inventories that include fewer species per site; the majority of species may remain undetected at each site (Taylor, 2002). These methods may be appropriate for broad observations of community patterns (Grytnes, 2003; Bahram et al., 2012) but may not be suited to examining range overlap among species between sites. Alternatively, intensive sampling methods (i.e., fewer sampling sites but more samples per site) tend to provide more comprehensive inventories of EM fungi at a given site. Intensive sampling may be necessary to demonstrate the presence or absence of range overlap and the role of the MDE.
In this study, we used intensive sampling to examine species range overlap and the possible role of the MDE in EM fungal richness along an elevation gradient on Mount Fuji, Japan. We also examined the relative importance of elevation and host tree species in explaining EM fungal composition along the gradient.
Materials and methods
Study sites and sampling
Field sampling was conducted on the northwest slope of Mt Fuji, central Japan, in 2011. This area has a temperate climate characterized by warm, wet summers and cool, moderately dry winters. We established four study sites in closed-canopy natural forests developed on a scoria deposit (Table 1; Miyamoto et al., 2013). These forests are estimated to be more than 300 years old and are among the ecosystems least affected by human activity. The highest sampling site (2250 m) was located just below tree line.
A complete list of tree species identified at the study sites is available in Miyamoto et al. (2013). Site 1 (1100 m) was dominated by Quercus crispula (36% of relative basal area), Fagus japonica (13%), Carpinus tschonoskii (10%) and Fagus crenata (8%). Site 2 (1550 m) was a deciduous–conifer mixed stand, dominated by F. crenata (47%), Abies homolepsis (29%) and Tilia japonica (12%). Sasamorpha borealis was the predominant groundcover at Site 2. The two higher-elevation sites (Sites 3 and 4) were characterized as subalpine coniferous forests exclusively dominated by Pinaceae species. Dominant trees at Site 3 (1900 m) included Abies veitchii (50%) and Tsuga diversifolia (44%); T. diversifolia (74%), Larix kaempferi (23%) and A. veitchii (3%) were the main species at Site 4 (2250 m). The tree species composition along the elevation gradient comprises typical vegetation in central Japan.
Fifty soil cores (5 × 5 cm to 10 cm deep) were collected from a 1-ha area at each site. A distance of 5–10 m was maintained between soil-sampling locations to ensure independence of the samples (Lilleskov et al., 2004; Pickles et al., 2009). Soil cores were stored separately in plastic bags at 4 °C until processing. A vegetation survey was conducted at every other soil-sampling location. Tree species and diameter at breast height (1.3 m) were recorded for all living trees (>1.3 m tall) within a 5-m radius of the sampling point. Litter depth and geographic coordinates (Garmin 62S; Garmin International, Olathe, KS, USA) were recorded at each sampling point.
Molecular analyses
All roots were carefully removed from each soil sample. EM root tips were classified by their morphological characteristics, including surface color, texture of the mantle surface, emanating hyphae and rhizomorphs. Healthy EM root tips (n=1–3) were sampled from each morphotype of each core for molecular analyses. Morphotyping was completed within 3 weeks after soil sampling.
We followed Murata et al. (2013), with minor modifications, for DNA extraction and molecular analyses. Briefly, fungal DNA was extracted from root tips using the cetyltrimethyl ammonium bromide method. Polymerase chain reaction was performed to amplify internal transcribed spacer (ITS) regions (ITS1, 5.8S, ITS2) of the rDNA using the forward primer ITS1F and various reverse primers, that is, ITS4, ITS4B, LR21, LR22 and LR5, depending on their amplification success with each morphotype. The amplified polymerase chain reaction products were checked on 1.2% agarose gels (0.5 × TBE buffer) and visualized under UV light to examine the quality and quantity of amplicons. Restriction fragment length polymorphism (RFLP) patterns were compared among replicates of each morphotype, and all polymerase chain reaction products with unique RFLP types in each morphotype were purified and sequenced. Sanger sequencing was conducted primarily using primer ITS1. Another sequencing primer, that is, ITS4, was additionally used for poorly sequenced samples.
High-quality ITS sequences >350 bp in length were aligned, manually edited and clustered into molecular operational taxonomic units (hereafter referred to as ‘species’) at ⩾97% similarity (Izzo et al., 2005; Murata et al., 2013) using ATGC ver. 7 (Genetyx Corp., Tokyo, Japan). The sequences were compared with known sequences in the international nucleotide sequence database; species names were assigned based on the taxonomy of those with which they shared the highest homology in the BLAST searches. Sequences that showed high homology with saprophytic and parasitic fungi or with non-fungal sequences were excluded from subsequent analyses. Cenococcum geophilum (hereafter referred to as ‘Cg’) is a species complex that is not well classified solely by variability in ITS regions (Douhan and Rizzo, 2005). Thus, Cg was identified based primarily on its unique morphology, as in many previous studies (e.g. Twieg et al., 2007; Murata et al., 2013).
Host trees associated with EM fungal species were identified to the genus by amplifying the trnL intron of chloroplast rDNA using primer pairs that amplified chloroplast trnC–trnD and trnE–trnF (Taberlet et al., 1991; Murata et al., 2013). RFLP patterns of root-tip samples were then compared with those obtained from leaves of host species identified in the field. Direct sequencing was applied for samples with unclear RFLP patterns. We did not treat Tilia as an EM host species because Tilia was not detected among the EM roots examined.
Soil analyses
Soil samples were air dried and passed through a 1-mm sieve, and the pH of each sample was measured using an aqueous slurry and HM-25G glass electrode (TOA DKK Corp., Tokyo, Japan). The soils were further sieved through a 250-μm screen and used to measure total C and total N by dynamic flash combustion with a Flash EA 1112 CN Analyzer (AMCO Inc., Tokyo, Japan).
Statistical analyses
Estimated species richness was computed for each site using Chao2 nonparametric estimators (Chao, 1984) and for the whole region using EstimateS ver. 8.20 (Colwell et al., 2012) with 1000 randomizations without replacement.
We tested whether the MDE models could explain observed richness patterns in EM fungi. We used a fully-stochastic null model that uses empirical data and places all species ranges entirely within the bounded domain (Colwell and Hurtt, 1997; Colwell and Lees, 2000). Mid-domain null simulations were conducted according to McCain (2004). The domain limits (1100–2250 m) and the number of divisions within the domain (n=4) were defined based on our sampling scale. Range sizes of recorded fungal species were calculated from the upper and lower limits of their occurrence. A Monte Carlo method was then used to compute estimated richness using randomly chosen range midpoints and empirical range sizes without replacement. Confidence limits (95%) were computed based on 50 000 simulations.
Simple linear and quadratic regression analyses were used to examine relationships between EM fungal richness and environmental factors. Environmental factors included mean annual temperature, mean total precipitation, litter depth, soil pH, soil C/N and richness of host-tree genera. Residuals of EM fungal richness from MDE were calculated by subtracting observed richness from the estimated richness computed by the mid-domain null model. The residuals were then used for regression analyses with environmental factors.
The occurrence of fungal species per host genus per site was treated as a sample unit. Hellinger transformation was applied to the community matrix to downweight the influence of rare species (Legendre and Gallagher, 2001). Community dissimilarities were visualized using non-metric multidimensional scaling on Bray–Curtis distances (999 permutations). The Mantel test was used to assess the spatial dependence of fungal species composition. Euclidean and Bray–Curtis distances were used for elevation and fungal species composition, respectively. Differences in EM fungal composition among host-tree genera and families were tested using a permutation-based multivariate analysis of variance test (ADONIS test, Vegan package in R). Bray–Curtis distances were used, and significance was tested with 999 permutations. The relative importance of elevation and host to fungal community composition was then analyzed using variation partitioning by redundancy analyses. These statistical analyses were conducted in the R Language and Environment for Statistical Computing (ver. 2.15). Statistical significance was defined as P<0.05, unless otherwise noted.
Results
General descriptions of EM fungal communities
Tree roots colonized by EM fungi were found in 197 of the 200 cores. DNA was extracted from 2813 root tips, and 1014 of 1347 sequenced tips (75%) were successfully identified (>350 bp). Eleven sequences did not belong to EM fungal taxa. In total, 85% of the morphotypes were successfully classified to species. We identified 302 EM fungal species, including 148 singletons (49.0% of the total) and 63 doubletons (20.9%) (Supplementary Table S1). Chao2-estimated richness across the entire study area was 475±38.3. Rarefaction curves of observed richness did not reach an asymptote at any site, whereas estimated (Chao2) richness became nearly asymptotic at all sites (Supplementary Figure S1).
Major lineages recorded in this study included: /tomentella-thelephora (59 species), /cortinarius (51), /russula-lactarius (42), /sebacina (30) and /inocybe (28). Most species were Basidiomycetes; Ascomycetes (12 species) were a minor component (4%). Cg was the most abundant fungal taxon, recorded in 156 of 200 soil cores (78.0%), followed by Russula bicolor sp.1 (20 cores, 10%) and Xerocomus sp.2 (17 cores, 8.5%). Cg occurred at all four sites. In the following sections, we present results obtained by excluding Cg from the analyses to avoid the influence of this incomparably abundant species complex and to improve analytical accuracy (some results including Cg are available in Supplementary Figure S2 and Table S2).
Elevation patterns
Both observed and estimated EM fungal richness showed a unimodal pattern, with the highest richness at 1550 m (Table 2 and Figure 1). The observed EM fungal richness–elevation pattern fit the 95% prediction curves computed from 50 000 simulations of the mid-domain null model (Figure 1). No climate or soil factors showed significant linear or quadratic correlation with observed fungal richness (P>0.1). However, residuals of EM fungal richness from the mid-domain null model were correlated with elevation (F1,2=14.2, R2=0.82, P=0.063), precipitation (F1,2=103.6, R2=0.97, P=0.009), soil C/N (F1,2=69.9, R2=0.96, P=0.014) and richness of host genera (F1,2=19.0, R2=0.86, P=0.049).
Observed richness (solid line) and the 95% confidence limits computed by the mid-domain null model based on 50 000 simulations (dotted lines). Error bars indicate the 95% confidence interval of observed richness, computed by EstimateS. Quadratic regression models were fitted to the richness and confidence limits.
The relative occurrence of each EM fungal lineage in a given site was defined as the number of cores in which a given lineage occurred divided by the total number of cores containing EM-colonized roots. Higher elevations were associated with higher relative occurrence of /cortinarius (6.9%, 7.6%, 20.8%, and 18.7%, respectively, from low- to high-elevation sites) and /russula-lactarius (15.4%, 6.9%, 20.0%, 24.2%). Lower-elevation sites showed higher relative occurrence of /sebacina (12.8%, 11.5%, 2.0%, 2.3%) and /inocybe (8.5%, 7.6%, 2.3%, 2.7%). The relative occurrence of /tomentella-thelephora (16.0%, 18.0%, 14.6%, 5.0%) showed a unimodal pattern along the elevation gradient, with the highest occurrence at 1550 m. Fisher’s exact tests showed that the relative occurrence of each of the above lineages significantly differed among sites (P<0.002).
Seventy-three (24.2%) species occurred in two or more sites. Piloderma fallax sp.2 and Sebacina sp.1 occurred in three sites (1550–2250 m). Most species that were present in >1 site (72 species) occurred in adjacent sites, and only one (Amphinema byssoides) appeared in non-adjacent sites (Figure 2). The lowest- and highest-elevation sites did not share any species in common. The χ2-test with Yates correction showed that the proportion of species shared among sites was significantly higher in adjacent site pairs than in non-adjacent site pairs (P<0.001).
Occurrence overlap of ectomycorrhizal fungal species across sites. (a) Cumulative number of fungal species, from Sites 1 to 4. Values in the columns are the numbers of species. Open columns indicate site-specific species and closed columns indicate site-shared species between adjacent site pairs. Shaded columns at the right end indicate two species that were found across three adjacent sites and one species found between non-adjacent sites (Sites 2 and 4). (b) Frequency of occurrence in each site. Columns correspond to (a), but values in the columns are relative frequency of occurrence in each site (values for the short columns are presented beside the columns or not shown for clarity). X axis is the cumulative frequency across sites. Supplementary Table S1 provides detailed information on species taxa and occurrence. Cg is excluded.
Effects of elevation and host species on EM fungal composition
Host genus was identified for 83% of the EM root tips. We excluded hosts represented by <5 soil cores, thus retaining 266 of the 301 EM fungal species (88.1%; excluding Cg) for the subsequent analyses.
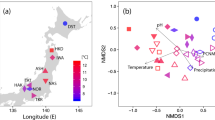
The Mantel test showed that dissimilarity in EM fungal composition increased with elevation (r=0.66, P<0.001). The non-metric multidimensional scaling plot revealed a separation of EM composition by site and host (Figure 3). Host effects were not significant at the genus level (F7,6=1.24, P=0.079) but were significant at the family level (F2,11=1.77, R2=0.24, P=0.009) based on ADONIS tests. Variation partitioning was applied to examine the relative importance of host family and elevation in explaining the variation in EM species composition. Elevation alone explained more variation (R2adj=0.026) than did host alone (not significant); elevation and host collectively explained the largest variance (R2adj=0.075) in fungal composition.
Non-metric multidimensional scaling plot showing similarity in EM fungal composition by site and host. Hosts represented by <5 soil samples were excluded from the analyses. Stress=0.017. Open circles represent Pinaceae, gray circles represent Fagaceae and black circles represent Betulaceae. Host genera are Fagus (F), Quercus (Q), Betula (B), Carpinus (C), Abies (A), Tsuga (T), Larix (L) and Pinus (P). The number following each genus indicates the sites.
The majority (42 of 53) of fungal species found in >1 site occurred on multiple host genera (Supplementary Table S3). We examined whether the distribution of site-shared species was affected by host genera by using variation partitioning to separate the effects of host and elevation. The results showed an insignificant effect of host genera and a significant effect of elevation (P=0.001, R2adj=0.201).
Discussion
We revealed high species richness of EM fungi on a slope of Mt Fuji, identifying 302 species and estimating that nearly 480 species exist in this location. This is among the highest richness reported from a single mountain area (Morris et al., 2008; Tedersoo et al., 2008; Bahram et al., 2012; Murata et al., 2013).
The MDE on species richness is recognized in many terrestrial macroorganisms (Colwell et al., 2004; McCain, 2005; Rahbek, 2005; Lee et al., 2013) and in freshwater macroinvertebrates (Wang et al., 2011), but this study is the first to demonstrate this effect in microorganisms and EM fungi. MDE models predict that random placement of species ranges within a bounded geographical domain leads to increased range overlap toward the center of the domain, producing a peak in species richness in that area (Colwell and Lees, 2000). The MDE models assume that species have continuous distribution ranges (Colwell et al., 2004; Currie and Kerr, 2008) that are potentially related to environmental gradients. Our results support the MDE in EM fungal richness. First, 99% (72 of 73) of fungal species found at two or more sites occurred at adjacent sites, showing that individual EM fungal species were distributed continuously along the gradient. Second, these continuous species distributions led to greater range overlap at mid-elevation than at higher and lower elevations (Figure 2). Finally, richness peaked in the mid-elevation site, and the pattern fit the 95% confidence limits computed by mid-domain null simulations (Figure 1). In our study area, the natural tree line was located just above the highest site (2250 m). Extensive residential areas and vegetation not associated with EM fungi extended below the lowest site (1100 m). Thus, the examined slope was geographically constrained, and EM fungi were unable to colonize habitats outside the boundaries of the study. This geographic setting and our empirical data suggest that geometric constraints are potential drivers of EM species richness patterns along elevation gradients.
We also analyzed deviation of the observed species richness from the mean values of the mid-domain null model. The deviation showed a negative correlation with elevation (P=0.063), indicating that unexplained richness patterns could be attributed to elevation-related factors. Indeed, the deviation was correlated with precipitation, soil (C/N) and richness of host-tree genera. These factors are not mutually exclusive and are likely to affect EM fungal richness via interaction effects (Bahram et al., 2012; Tedersoo et al., 2012). Our results suggest that the MDE can be an effective null model for isolating significant environmental factors that determine microbial richness.
Global meta-analyses suggest that host family is the main driver of fungal community composition (Tedersoo et al., 2012). However, importance of hosts tends to vary by host groups, regions and scales (Smith et al., 2009; Tedersoo et al., 2011; Murata et al., 2013). Host effects are also less clear when the distribution of host is determined by geographic position and environmental conditions. In our study sites, various host trees were distributed along the elevation gradient within a narrow geographical area, enabling us to separate the effects of elevation and host on EM species composition.
Variation partitioning showed that elevation explained more of the variability in EM fungal composition than did host. Many studies have reported strong host effects on EM fungal communities at the stand scale (Ishida et al., 2007; Morris et al., 2008; Smith et al., 2009), but other factors (e.g., climate) may be more important at larger scales (Lilleskov and Parrent, 2007; Bahram et al., 2012). For example, the EM composition of co-occurring Fagus and Abies spp. in a mixed stand was more similar than that of Abies pairs in adjacent conifer stands (Figure 3). The same host genera in the two deciduous stands also showed clear separations in fungal composition by elevation. These results may indicate that many EM fungi have evolved compatibility to a wide range of co-occurring host lineages, such as Pinaceae, Fagaceae and Betulaceae that share overlapping geographic ranges (Bahram et al., 2013). Several studies also have suggested that host shifts among EM fungi prevail when preferable hosts are absent within a community (Trocha et al., 2012; Wolfe and Pringle, 2012; Bahram et al., 2013; Murata et al., 2013).
It should be noted that the interaction effect of elevation and host explained more of the variance in EM species composition than did either elevation or host alone. Furthermore, two conifer stands at the two higher-elevation sites shared many common EM fungi despite the differences in elevation. These results reflected the interaction effects and varying importance of elevation and host species on the composition of EM fungal communities.
Conclusions
Microbial communities are usually represented by numerous rare species, which are inevitably overlooked with lower sampling efforts. Thus, sampled microbial communities may not represent the existing microbial communities in the field, often making it inappropriate to apply ecological models originally developed for macroorganisms to microorganisms. In particular, demonstrating geographical distributions of microbes is challenging. In this study, we used an intensive sampling approach and obtained relatively large EM fungal community data at each site. This approach enabled us to detect many site-shared species across the sites along the elevation gradient. Interestingly, these site-shared species nearly always appeared in adjacent sites along the gradient, clearly confirming the existence of species distribution range along the gradient. Furthermore, observed EM fungal richness was within the range of estimated richness under the MDE null model, which is based on the assumption that individual species have geographical distribution ranges. These findings indicate that intensive sampling approaches are promising in ecological studies on microbes. Although we used a single gradient in this study because of the tradeoff between sampling intensity and number of sites, further investigations of microbial community patterns by intensive approaches in other gradients and meta-analyses would confirm the generality of our findings.
References
Aponte C, Garcia LV, Maranon T, Gardes M . (2010). Indirect host effect on ectomycorrhizal fungi: leaf fall and litter quality explain changes in fungal communities on the roots of co-occurring Mediterranean oaks. Soil Biol Biochem 42: 788–796.
Bahram M, Polme S, Koljalg U, Zarre S, Tedersoo L . (2012). Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytol 193: 465–473.
Bahram M, Koljalg U, Kohout P, Mirshahvaladi S, Tedersoo L . (2013). Ectomycorrhizal fungi of exotic pine plantations in relation to native host trees in Iran: evidence of host range expansion by local symbionts to distantly related host taxa. Mycorrhiza 23: 11–19.
Chao A . (1984). Non-parametric estimation of the number of classes in a population. Scand J Statist 11: 265–270.
Colwell RK, Hurtt GC . (1997). Nonbiological gradients in species richness and a spurious Rapoport effect. Am Nat 144: 570–595.
Colwell RK, Lees DC . (2000). The mid-domain effect: geometric constraints on the geography of species richness. Trends Ecol Evol 15: 70–76.
Colwell RK, Rahbek C, Gotelli NJ . (2004). The mid-domain effect and species richness patterns: What have we learned so far? Am Nat 163: E1–E23.
Colwell RK, Chao A, Gotelli NJ, Lin S-Y, Mao CX, Chazdon RL et al. (2012). Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J Plant Ecol 5: 3–21.
Courty P-E, Buee M, Diedhiou AG, Frey-Klett P, Le Tacon F, Rineau F et al. (2010). The role of ectomycorrhizal communities in forest ecosystem processes: new perspectives and emerging concepts. Soil Biol Biochem 42: 679–698.
Cox F, Barsoum N, Lilleskov EA, Bidartondo MI . (2010). Nitrogen availability is a primary determinant of conifer mycorrhizas across complex environmental gradients. Ecol Lett 13: 1103–1113.
Currie DJ, Kerr JT . (2008). Tests of the mid-domain hypothesis: a review of the evidence. Ecol Monogr 78: 3–18.
Douhan GW, Rizzo DM . (2005). Phylogenetic divergence in a local population of the ectomycorrhizal fungus Cenococcum geophilum. New Phytol 166: 263–271.
Gaston KJ . (2000). Global patterns in biodiversity. Nature 405: 220–227.
Grytnes JA . (2003). Species-richness patterns of vascular plants along seven altitudinal transects in Norway. Ecography 26: 291–300.
Hillebrand H . (2004). On the generality of the latitudinal diversity gradient. Am Nat 163: 192–211.
Hobbie EA . (2006). Carbon allocation to ectomycorrhizal fungi correlates with belowground allocation in culture studies. Ecology 87: 563–569.
Hobbie JE, Hobbie EA . (2006). N-15 in symbiotic fungi and plants estimates nitrogen and carbon flux rates in Arctic tundra. Ecology 87: 816–822.
Horton TR, Bruns TD, Parker VT . (1999). Ectomycorrhizal fungi associated with Arctostaphylos contribute to Pseudotsuga menziesii establishment. Can J Bot 77: 93–102.
Horton TR, Bruns TD . (2001). The molecular revolution in ectomycorrhizal ecology: peeking into the black-box. Mol Ecol 10: 1855–1871.
Ishida TA, Nara K, Hogetsu T . (2007). Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broadleaf forests. New Phytol 174: 430–440.
Izzo A, Agbowo J, Bruns TD . (2005). Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. New Phytol 166: 619–630.
The Japan Meteorological Agency. (2011) Mesh Average 2010. Japan Meteorological Business Support Center: Tokyo, Japan.
Kluge J, Kessler M, Dunn RR . (2006). What drives elevational patterns of diversity? A test of geometric constraints, climate and species pool effects for pteridophytes on an elevational gradient in Costa Rica. Global Ecol Biogeogr 15: 358–371.
Koide RT, Fernandez C, Petprakob K . (2011). General principles in the community ecology of ectomycorrhizal fungi. Ann For Sci 68: 45–55.
Lee CB, Chun JH, Song HK, Cho HJ . (2013). Altitudinal patterns of plant species richness on the Baekdudaegan Mountains, South Korea: mid-domain effect, area, climate, and Rapoport's rule. Ecol Res 28: 67–79.
Legendre P, Gallagher ED . (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129: 271–280.
Lilleskov EA, Bruns TD, Horton TR, Taylor DL, Grogan P . (2004). Detection of forest stand-level spatial structure in ectomycorrhizal fungal communities. FEMS Microbiol Ecol 49: 319–332.
Lilleskov EA, Parrent JL . (2007). Can we develop general predictive models of mycorrhizal fungal community-environment relationships? New Phytol 174: 250–256.
McCain CM . (2004). The mid-domain effect applied to elevational gradients: species richness of small mammals in Costa Rica. J Biogeogr 31: 19–31.
McCain CM . (2005). Elevational gradients in diversity of small mammals. Ecology 86: 366–372.
McCain CM . (2009). Vertebrate range sizes indicate that mountains may be 'higher' in the tropics. Ecol Lett 12: 550–560.
Miyamoto Y, Nakano T, Nara K . (2013). Diversity and composition of tree species in mature stands along an altitudinal gradient on Mt. Fuji. Mount Fuji Research 7: 19–22.
Morris MH, Smith ME, Rizzo DM, Rejmanek M, Bledsoe CS . (2008). Contrasting ectomycorrhizal fungal communities on the roots of co-occurring oaks (Quercus spp.) in a California woodland. New Phytol 178: 167–176.
Murata M, Kinoshita A, Nara K . (2013). Revisiting the host effect on ectomycorrhizal fungal communities: implications from host–fungal associations in relict Pseudotsuga japonica forests. Mycorrhiza 23: 641–653.
Nara K, Nakaya H, Wu BY, Zhou ZH, Hogetsu T . (2003). Underground primary succession of ectomycorrhizal fungi in a volcanic desert on Mount Fuji. New Phytol 159: 743–756.
Nara K . (2006). Ectomycorrhizal networks and seedling establishment during early primary succession. New Phytol 169: 169–178.
Peay KG, Kennedy PG, Bruns TD . (2008). Fungal community ecology: a hybrid beast with a molecular master. Bioscience 58: 799–810.
Peay KG, Kennedy PG, Davies SJ, Tan S, Bruns TD . (2010). Potential link between plant and fungal distributions in a dipterocarp rainforest: community and phylogenetic structure of tropical ectomycorrhizal fungi across a plant and soil ecotone. New Phytol 185: 529–542.
Pickles BJ, Genney D, Anderson IC, Alexander IJ . (2009). Spatial ecology of ectomycorrhizas: analytical strategies. In: Azcon-Aguilar C, Barea JM, Gianinazzi S, Gianinazzi-Pearson V, (eds) Mycorrhizas. Functional Processes and Ecological Impact. Springer: Berlin, Germany, pp 155–165.
Pickles BJ, Genney DR, Anderson IC, Alexander IJ . (2012). Spatial analysis of ectomycorrhizal fungi reveals that root tip communities are structured by competitive interactions. Mol Ecol 21: 5110–5123.
Rahbek C . (2005). The role of spatial scale and the perception of large-scale species-richness patterns. Ecol Lett 8: 224–239.
Rineau F, Courty P-E . (2011). Secreted enzymatic activities of ectomycorrhizal fungi as a case study of functional diversity and functional redundancy. Ann For Sci 68: 69–80.
Smith ME, Douhan GW, Fremier AK, Rizzo DM . (2009). Are true multihost fungi the exception or the rule? Dominant ectomycorrhizal fungi on Pinus sabiniana differ from those on co-occurring Quercus species. New Phytol 182: 295–299.
Taberlet P, Gielly L, Pautou G, Bouvet J . (1991). Universal primers for amplification of 3 noncoding regions of chloroplast DNA. Plant Mol Biol 17: 1105–1109.
Taylor AFS . (2002). Fungal diversity in ectomycorrhizal communities: sampling effort and species detection. Plant Soil 244: 19–28.
Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I et al. (2008). Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol 180: 479–490.
Tedersoo L, May TW, Smith ME . (2010). Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20: 217–263.
Tedersoo L, Bahram M, Jairus T, Bechem E, Chinoya S, Mpumba R et al. (2011). Spatial structure and the effects of host and soil environments on communities of ectomycorrhizal fungi in wooded savannas and rain forests of Continental Africa and Madagascar. Mol Ecol 20: 3071–3080.
Tedersoo L, Bahram M, Toots M, Diedhiou AG, Henkel TW, Kjoller R et al. (2012). Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol Ecol 21: 4160–4170.
Trocha LK, Kalucka I, Stasinska M, Nowak W, Dabert M, Leski T et al. (2012). Ectomycorrhizal fungal communities of native and non-native Pinus and Quercus species in a common garden of 35-year-old trees. Mycorrhiza 22: 121–134.
Twieg BD, Durall DM, Simard SW . (2007). Ectomycorrhizal fungal succession in mixed temperate forests. New Phytol 176: 437–447.
Visser S . (1995). Ectomycorrhizal fungal succession in Jack pine stands following wildfire. New Phytol 129: 389–401.
Wang J, Soininen J, Zhang Y, Wang B, Yang X, Shen J . (2011). Contrasting patterns in elevational diversity between microorganisms and macroorganisms. J Biogeogr 38: 595–603.
Wolfe BE, Pringle A . (2012). Geographically structured host specificity is caused by the range expansions and host shifts of a symbiotic fungus. ISME J 6: 745–755.
Acknowledgements
This project was funded by the Grants-in-Aid for JSPS Fellows to YM and Grants-in-Aid for Scientific Research to KN. We thank Katsuaki Jimbo, Sho Ohashi, Kenshiro Minami and Dr Masao Murata for field assistance; Dr Akihiko Kinoshita and Dr Megumi Tanaka for lab assistance; Dr Kenshiro Oshima, Emi Omori and Yukiko Takayama for assistance of sequencing.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Miyamoto, Y., Nakano, T., Hattori, M. et al. The mid-domain effect in ectomycorrhizal fungi: range overlap along an elevation gradient on Mount Fuji, Japan. ISME J 8, 1739–1746 (2014). https://doi.org/10.1038/ismej.2014.34
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.34
Keywords
This article is cited by
-
Community composition of phytopathogenic fungi significantly influences ectomycorrhizal fungal communities during subtropical forest succession
Applied Microbiology and Biotechnology (2024)
-
Species composition of root-associated mycobiome of ruderal invasive Anthemis cotula L. varies with elevation in Kashmir Himalaya
International Microbiology (2023)
-
The diversity of moths (Erebidae: Arctiinae: Arctiini) from threatened mountain cloud forests in the Mesoamerican biodiversity hotspot
Journal of Insect Conservation (2023)
-
Diversity and distribution of CO2-fixing microbial community along elevation gradients in meadow soils on the Tibetan Plateau
Scientific Reports (2022)
-
Temperature and Precipitation Drive Elevational Patterns of Microbial Beta Diversity in Alpine Grasslands
Microbial Ecology (2022)