Abstract
Marine photosynthesis is one of the major contributors to the global carbon cycle and the world’s oxygen supply. This process is largely driven by cyanobacteria, namely Synechococcus and Prochlorococcus. Genes encoding photosystem-II (PSII) reaction center proteins are found in many cyanophage genomes, and are expressed during the infection of their hosts. On the basis of metagenomics, cyanophage photosystem-I (PSI) gene cassettes were recently discovered with two gene arrangements psaJF→C→A→B→K→E→D and psaD→C→A→B. It was suggested that the horizontal transfer of PSII and PSI genes is increasing phage fitness. To better understand their diversity, we designed degenerate primers to cover a wide diversity of organisms, and using PCR we targeted the psaC→A arrangement, which is unique to cyanophages cassettes. We examined viral concentrates from four islands in the Pacific Ocean and found samples containing the psaC→A arrangement. Analyses of the amplified viral psaA gene revealed six subgroups varying in their level of similarity and %G+C content, suggesting that the diversity of cyanophage PSI genes is greater than originally thought.
Similar content being viewed by others
Introduction
Marine cyanobacteria of the Synechococcus and Prochlorococcus genera are significant contributors to global photosynthesis (Li et al., 1993; Liu et al., 1997; Partensky et al., 1999). It was shown that a number of viruses (cyanophages) that infect these cyanobacteria carry photosynthetic genes coding for photosystem-II (PSII) proteins (Mann et al., 2003; Lindell et al., 2004; Millard et al., 2004; Lindell et al., 2005; Sullivan et al., 2005; Sullivan et al., 2006; Sharon et al., 2007) and photosystem-I (PSI) proteins (Sharon et al., 2009; Alperovitch-Lavy et al., 2011; Béjà et al., 2012).
Three viral PSI gene organizations are currently known: a single psaJ gene (Sharon et al., 2011), and two gene cassettes psaJF→C→A→B→K→E→D and psaD→C→A→B (Sharon et al., 2009; Alperovitch-Lavy et al., 2011; Béjà et al., 2012). Although the longer cassette is readily detected in more than 20 Global Ocean Sampling expedition (GOS) (Rusch et al., 2007)) scaffolds (Sharon et al., 2009; Alperovitch-Lavy et al., 2011; Béjà et al., 2012)], fragments from the short cassette were detected only three times in different metagenomic data sets (Béjà et al., 2012). In addition to differences at the protein level, psaA genes coming from the psaD→C→A→B gene organization are distinguishable in their higher %G+C content compared with psaA genes coming from the long viral gene organization (∼50%G+C and ∼40%G+C, respectively; Figure 1).
Schematic physical maps of viral long PCR, viral GOS sequences and cyanobacteria containing PSI gene cassettes. PSI genes are colored according to their %G+C content. Gray arrows represent viral open reading frames. Color code indexes indicate %G+C; the calculations were performed for each gene separately. Positions of forward and reverse primers are indicated by black arrows. A phage symbol is attached to each sequence identified as also containing structural viral genes. For space considerations, cyanobacterial gene arrangements are shown only for Prochlorococcus Med4 and Synechococcus WH8109. The arrangement psaC gene followed immediately by a psaA gene was not detected in any of the currently available cyanobacterial genomes (37 species in CyanoBase (http://genome.microbedb.jp/cyanobase); November 2014).
Cyanophages containing the psaJF→C→A→B→K→E→D gene organization were hypothesized to produce a monomeric PSI (as opposed to a trimeric PSI observed in their cyanobacterial hosts and similarly to what is observed in plants) and to shift host metabolism toward a cyclic photosynthetic mode (Sharon et al., 2009; Philosof et al., 2011). Therefore, infected cells are assumed to keep on harvesting light energy while avoiding the fixation of CO2. Hence, the gained energy (in the form of ATP) is used to enhance nucleotide biosynthesis, required for the replication of additional viral genomes (Sharon et al., 2009; Philosof et al., 2011; Thompson et al., 2011; Enav et al., 2014). Current data about the psaD→C→A→B arrangement is so scarce that no working hypothesis have been put forward yet beside the mentioning that an ancestral PSI composed of only these four subunits is a plausible scenario (Nelson, 2011).
The gene combination psaC→psaA is present in both the long and short viral cassettes, however, it is absent in their potential cyanobacterial marine hosts Prochlorococcus and Synechococcus (Figure 1). Therefore, in order to better understand the diversity of viral PSI genes, we used general degenerate primers designed against PsaC and PsaA proteins to amplify specifically viral psaC→psaA amplicons from different environmental samples.
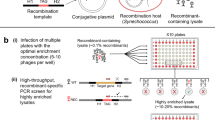
Materials and methods
PsaC-PsaA gene amplification and cloning
Degenerate primers were designed against the PSI PsaC and PsaA proteins (Primers PsaCdeg-fwd, PsaAdeg2-rev and PsaA-rev with 64 128 and 256 degeneracy, respectively; see Supplementary Table S1 for primer sequences) based on the multiple sequence alignment of cyanobacterial and viral proteins obtained from the GOS data set (Rusch et al., 2007) and viral PSI proteins previously discovered (Sharon et al., 2009; Béjà et al., 2012). PCR reactions using primers to target the psaC→A gene arrangement (Figure 1; each reaction contained a forward primer and one of the reverse primers) were performed directly on viral concentrates from the Pacific Southern Line Islands (collected in April 2009 from the Millennium, Flint, Malden and Starbuck Islands) (Supplementary Figure S1; see Supplementary Tables S1 and 2 in Kelly et al. (2014) for metadata details on samples Millennium 9, Flint 6, Malden5 and Starbuck 7). Viral concentrates were prepared according to Dinsdale et al. (2008). Each PCR reaction was performed using BIO-X-ACTShort mix (Bioline, London, UK). PCR amplification was carried out in a total volume of 30 μl containing 1 μl of phage concentrate as template, 0.8 mM dNTPs, 2 mM MgCl2, 1 μM primers (each) and 2.4 U BIO-X-ACTShort DNA polymerase. The amplification conditions included the steps at 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 1 min. PCR products (450–530 bp with the inner reverse primer and 850 bp with the outer reverse primer) were subcloned using the QIAGEN (Hilden, Germany) PCR cloning kit according to the manufacturer’s specifications and sequenced using Sanger sequencing (Macrogen Europe, Amsterdam, NL, USA).
Tetranucleotide frequency analysis
In order to profile the genomic composition of different psaA genes, each psaA sequence was represented by a tetranucleotide frequency vector (256 features per vector). Frequencies were calculated exclusively from the coding strand using overlapping windows (window location was modified by single-nucleotide steps) and normalized to the sequence length (See Supplementary file S1 for the alignment).
To discover possible differences between the genomic compositions of psaA genes, principal component analysis was performed using tetranucleotide frequency vectors as the input value matrix. The three most variable principal components were plotted using GNU R RGL and ColorRamp packages (Keitt, 2008).
To better define the similarities and differences between the groups of psaA genes we calculated the cosine distance for each of the two genomic profiles (equation 1). We generated a symmetrical distance matrix of cosine similarities between all the genomic profiles and clustered the matrix using MultiExperiment Viewer (MeV) software (Saeed et al., 2006; hierarchical clustering method set to Pearson’s correlation, average linkage).

PsaA phylogenetic tree construction and analysis
PCR products obtained were trimmed to include only the psaA gene portion (PsaC proteins are too conserved to be used as phylogenetic markers) and translated according to the correct open reading frame. PsaA protein sequences from the PCR products and reference sequences from the GOS project (downloaded from the CAMERA website (http://camera.crbs.ucsd.edu/projects/)), and various Prochlorococcus and Synechococcus (downloaded from GenBank) were then aligned using Muscle under default parameters (Edgar, 2004). Maximum-likelihood phylogenetic trees were constructed using the phylogeny.fr pipeline (Dereeper et al., 2008), with PhyML v 3.0 (Guindon et al., 2010) and the WAG substitution model for amino acids (Whelan and Goldman, 2001). Five hundred bootstrap replicates were conducted for each analysis. Trees were visualized using FigTree v 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). See Supplementary file S2 for the alignment used to construct the tree.
Analyses of Southern Line Islands microbiomes
The microbial metagenomes of the samples used in this study for PCR have been previously deposited in the MG-RAST Metagenomics Analysis Server (Meyer et al., 2008), http://metagenomics.anl.gov/linkin.cgi?project=9220 (project name: Pacific Reef Microbiomes; Kelly et al., 2014). MG-RAST automated platform analyses were used in order to calculate the proportions of different cyanobacterial groups in these stations.
Shannon entropy
Shannon entropy was calculated using the Shannon Entropy-One tool from the HIV database (http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy_one.html). Reported values are summation of the position-specific values outputted by the tool.
Results and Discussion
The PsaC forward primer designed based on a conserved domain at the C-terminus of the protein and two reverse primers designed against the PsaA protein were used in order to perform PCR reactions that will amplify viral-only gene arrangements (Figure 1 and Supplementary Table S1). Using these primers, PCR was performed directly on viral concentrates collected from the remote Pacific Line Islands (see Map in Supplementary Figure S1), where high proportions of cyanophages containing PSI genes were reported (Sharon et al., 2009). Tetranucleotide frequency analysis of the amplified environmental viral psaA genes (that is, the psaA gene from the psaCA amplicon) revealed six viral psaA subgroups (Figure 2, and PCA movie S1). The optimal number of subgroups was determined using pseudo-F statistic with 100 iterations (Caliński and Harabasz, 1974). One group was composed of ∼50%G+C psaA amplicons whereas the other five groups contained ∼40%G+C psaA amplicons. Only 17% of the amplified psaA sequences were closely similar to the few previously reported GOS viral psaA sequences (⩾97% on the nucleotide level). Shannon entropy measures (Supplementary Table S2) show that the diversity of the viral high- and low-%G+C psaA groups is higher than what is observed in the Synechococcus and HL Prochlorococcus groups. Although the LL Prochlorococcus share the same ecological niche, the observed high Shannon entropy is a result of different %G+C observed within LL Prochlorococcus groups.
Distance matrix based on tetranucleotide frequencies of psaA gene sequences from GOS, Prochlorococcus, marine Synechococcus and PCR amplicones from the viral concentrates. Colors range from red, indicating high similarity, to blue, indicating higher distance. Black dots represent reported sequences from the GOS expedition. Synechococcus (Syn), Low Light adapted (LL) and High Light adapted (HL) Prochlorococcus (Pro) groups are labelled accordingly. Separation into different LL Prochlorococcus groups is according to Scanlan et al. (2009). Names were removed for clarity; see Supplementary Figure S2 for full names.
Overall, 12% of the clones were from the 50%G+C psaA cluster, however, the proportion markedly changed between the stations (Malden 25% (n=20), Millennium 19% (n=61), Starbuck 1.9% (n=51) and Flint 0% (n=7)). We speculate that this indicates on the availability of certain cyanobacterial hosts, which were present at the time of sampling in the Malden and Millennium Islands. Malden and Millennium islands had higher proportions of marine Synechococcus versus Prochlorococcus in the metagenomic data (73% Synechococcus, on the genus level), which might be explained by the higher proportions of nitrite and nitrate compared with ammonium observed in these stations (Moore et al., 2002; (see Supplementary Table S1 in Kelly et al.( 2014)). In contrast, different cyanobacterial proportions are observed in Starbuck and Flint islands (34% and 54% Synechococcus, respectively). However, we could not correlate any specific Synechococcus strains with the high-%G+C psaCA gene organization.
At the protein level, the environmental PsaA proteins were separated into two statistically supported clusters (Figure 3), with each cluster composed of PsaA proteins predicted to originate either from the psaJF →C→A→B→K→E→D or psaD→C→A→B gene organizations (predicted based on %G+C content of the corresponding genes). The separation into two distinct protein groups could either be the result of simply being operated in different hosts (that is, Prochlorococcus or marine Synechococcus), or could indicate operation at different photosynthetic modes in similar hosts (for example, working in a cyclic mode or in monomeric or trimeric PSI complexes (Sharon et al., 2009; Alperovitch-Lavy et al., 2011; Philosof et al., 2011)).
PsaA phylogenetic tree. Diamond symbols represent bootstrap values higher than 70%. Circle color represents %G+C classification according to the color index in Figure 1: green and yellow—High (∼50%), purple—Low (∼40%). Black dots represent sequences from the GOS expedition. A phage symbol is attached to each GOS sequence identified as also containing structural viral genes. The scale bar indicates the average number of amino-acid substitutions per site. Names were removed for clarity; see Supplementary Figure S3 for full names.
The use of PCR primers designed to amplify a specific gene arrangement enabled the screening for this unique viral psaCA gene combination without the need to clean the sample from other possible gene arrangements that might react with the primers, but are separated by a long DNA stretch or are in the wrong orientation (such as those found in various Prochlorococcus or marine Synechococcus). We have not ruled out the possibility that the unique psaCA gene organizations we obtained by PCR, originate from yet uncultured cyanobacteria. However, based on the existence of viral genes on the GOS scaffolds with similar gene organization and the amplification from viral concentrates, we find this scenario highly unlikely.
Our simple yet efficient targeted screen of viral photosynthetic genes revealed greater diversity of these genes than anticipated. Following the discovery of the single psaJ gene (Sharon et al., 2011) and the two currently known PSI gene cassettes (Sharon et al., 2009; Alperovitch-Lavy et al., 2011; Béjà et al., 2012), this new finding may suggest that other viral PSI gene arrangements and modes of photosynthetic operation are awaiting discovery.
References
Alperovitch-Lavy A, Sharon I, Rohwer F, Aro EM, Milo R, Nelson N et al. (2011). Reconstructing a puzzle: Existence of cyanophages containing both photosystem-I & photosystem-II gene-suites inferred from oceanic metagenomic datasets. Environ Microbiol 13: 24–32.
Béjà O, Fridman S, Glaser F . (2012). Viral clones from the GOS expedition with an unusual photosystem-I gene cassette organization. ISME J 6: 1617–1620.
Caliński T, Harabasz J . (1974). A dendrite method for cluster analysis. Commun Statist Theor Method 3: 1–27.
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469.
Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L et al. (2008). Microbial ecology of four coral atolls in the northern Line Islands. PLoS One 3: e1584.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Enav H, Mandel-Gutfreund Y, Béjà O . (2014). Comparative metagenomic analyses reveal viral-induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome 2: 9.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O . (2010). New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321.
Keitt TH . (2008). Coherent ecological dynamics induced by large scale disturbance. Nature 454: 331–334.
Kelly LW, Williams GJ, Barott KL, Carlson CA, Dinsdale EA, Edwards RA et al. (2014). Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc Natl Acad Sci USA 111: 10227–10232.
Li WKW, Zohary T, Yacobi YZ, Wood AM . (1993). Ultraphytoplankton in the eastern Mediterranean Sea-towards deriving phytoplankton biomass from flow cytometric measurements of abundance, fluorescence and light scatter. Mar Ecol Prog Ser 102: 79–87.
Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW . (2005). Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438: 86–89.
Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW . (2004). Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA 101: 11013–11018.
Liu H, Nolla HA, Campbell L . (1997). Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat Microb Ecol 12: 39–47.
Mann NH, Cook A, Millard A, Bailey S, Clokie M . (2003). Bacterial photosynthesis genes in a virus. Nature 424: 741.
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M et al. (2008). The Metagenomics RAST server - A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386.
Millard A, Clokie MRJ, Shub DA, Mann NH . (2004). Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA 101: 11007–11012.
Moore LR, Post AF, Rocap G, Chisholm SW . (2002). Utilization of different nitrogen sources by marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr 47: 989–996.
Nelson N . (2011). Photosystems and global effects of oxygenic photosynthesis. Biochim Biophys Acta 1807: 856–863.
Partensky F, Hess WR, Vaulot D . (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63: 106–127.
Philosof A, Battchikova N, Aro E-M, Béjà O . (2011). Marine cyanophages: Tinkering with the electron transport chain. ISME J 5: 1568–1570.
Rusch DB, Halpern AL, Heidelberg KB, Sutton G, Williamson SJ, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: I, The northwest Atlantic through the eastern tropical Pacific. PLoS Biol 5: e77.
Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA et al. (2006). TM4 microarray software suite. Methods Enzymol 411: 134–193.
Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299.
Sharon I, Battchikova N, Aro E-M, Giglione C, Meinnel T, Glaser F et al. (2011). Comparative metagenomics of microbial traits within oceanic viral communities. ISME J 5: 1178–1190.
Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N et al. (2009). Photosystem-I gene cassettes are present in marine virus genomes. Nature 461: 258–262.
Sharon I, Tzahor S, Williamson S, Shmoish M, Man-Aharonovich D, Rusch DB et al. (2007). Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J 1: 492–501.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW . (2005). Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol 3: e144.
Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW . (2006). Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol 4: e234.
Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J et al. (2011). Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA 108: e757–e764.
Whelan S, Goldman N . (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699.
Acknowledgements
We thank Ben Knowles for help with samples logistics and metadata and Michael Shmoish for help with statistics. This work was supported by a European Commission ERC Advanced Grant (no. 321647) and an ISF grant (no. 580/10). The psaC and psaA sequences reported here have been deposited with GenBank under accession numbers KF577926-KF577928 and KJ917402-KJ917544.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Hevroni, G., Enav, H., Rohwer, F. et al. Diversity of viral photosystem-I psaA genes. ISME J 9, 1892–1898 (2015). https://doi.org/10.1038/ismej.2014.244
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.244
This article is cited by
-
Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes
Microbiome (2020)
-
Prochlorococcus viruses—From biodiversity to biogeochemical cycles
Science China Earth Sciences (2018)
-
A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells
Nature Microbiology (2017)