Abstract
Cyanopodoviruses affect the mortality and population dynamics of the unicellular picocyanobacteria Prochlorococcus and Synechococcus, the dominant primary producers in the oceans. Known cyanopodoviruses all contain the DNA polymerase gene (DNA pol) that is important for phage DNA replication and widely used in field quantification and diversity studies. However, we isolated 18 cyanopodoviruses without identifiable DNA pol. They form a new MPP-C clade that was separated from the existing MPP-A, MPP-B, and P-RSP2 clades. The MPP-C phages have the smallest genomes (37.3–37.9 kb) among sequenced cyanophages, and show longer latent periods than the MPP-B phages. Metagenomic reads of both clades are highly abundant in surface waters, but the MPP-C phages show higher relative abundance in surface waters than in deeper waters, while MPP-B phages have higher relative abundance in deeper waters. Our study reveals that cyanophages with distinct genomic contents and infection kinetics can exhibit different depth profiles in the oceans.
Similar content being viewed by others
Introduction
Viruses are the most abundant biological entities in the oceans and play important roles in the marine ecosystem [1, 2]. Marine viruses can control microbial population dynamics through the lysis of host cells, mediate horizontal gene transfer among host genomes, and influence the marine biogeochemical cycle by expressing virus-encoded auxiliary metabolic genes during infection [3,4,5,6,7]. Due to the rapid developments in culture-independent approaches, especially viral metagenomics, our knowledge about the genetic diversity of marine viruses has increased considerably in recent years [8,9,10]. Despite a large number of newly discovered viral genomes by metagenomics, more than 90% of sequencing reads in many marine samples are derived from unknown viruses, indicating a need for culture-dependent efforts to further characterize viral dark matter [11].
One of the most important and widely distributed groups of marine viruses are cyanophages (viruses that infect cyanobacteria) [12,13,14]. Marine cyanobacteria, mainly of the genera Prochlorococcus and Synechococcus, are the most abundant photosynthetic organisms and responsible for more than 25% of primary production in the open ocean, contributing significantly to global CO2 fixation [15, 16]. In the ocean 1%–15% of marine cyanobacteria are infected by cyanophages [17, 18], and in some oceanic regions more than 46% of cyanobacteria can be killed by phages daily [19]. Via infection, cyanophages could greatly impact the population size, community dynamics [18, 20, 21], genome diversity and evolution of cyanobacteria [22, 23], as well as marine biogeochemical cycles [24,25,26,27,28]. Marine cyanophages isolated thus far include several distinct groups: cyanopodoviruses belonging to the T7-like group, cyanomyoviruses belonging to the T4-like or TIM5-like groups, and cyanosiphoviruses [29,30,31,32,33,34,35]. The T7-like cyanopodoviruses are frequently isolated and well represented in many oceanic metagenomic datasets, representing a ubiquitous and ecologically important viral component in the ocean [36,37,38].
The first sequenced marine cyanopodovirus genome (cyanophage P60 isolated from Synechococcus WH7805) contains the DNA polymerase gene (DNA pol) that is conserved in podoviruses infecting different heterotrophic bacteria [39, 40]. So far, DNA pol is present in all 18 published marine cyanopodovirus genomes [29, 34, 40,41,42] and 3 unpublished cyanopodovirus genomes in GenBank (Table 1). Based on the phylogenetic tree of DNA pol, marine cyanopodoviruses form a monophyletic MPP (Marine Picocyanobacteria Podovirus) subgroup that is further divided into MPP-A and MPP-B clusters [43], with the cyanophage P-RSP2 isolated from Prochlorococcus MIT9302 being an outlier [34].
Given its conservation in podovirus genomes and crucial function in phage genome replication, DNA pol has been widely used as a genetic marker for quantification and genetic diversity studies of cyanopodoviruses [7, 36, 37, 44,45,46]. However, evidence indicating the existence of cyanopodoviruses without DNA pol is accumulating. For example, the work examining the depth distribution of cyanopodoviruses revealed significantly lower abundances based on DNA pol than those based on the terminase gene in some samples [7]. The discrepancy may be due to the inability to recover all naturally occurring cyanopodoviruses by DNA pol. Furthermore, a polony method based on T4-like and T7-like cyanophage primers showed that the ratio of cyanophages to the total virus-like particles in the North Pacific Subtropical Gyre is 2–3 fold lower than that of cyanobacteria to the total bacteria, suggesting unknown cyanophage groups [45]. Likewise, unknown cyanophage types were found in different oceanic regions, and they could not be amplified by PCR primers targeting lineage-specific signature genes for the T4-like (portal protein gene g20), TIM5-like (DNA pol), and T7-like (DNA pol) cyanophages [47, 48].
Here we report a novel lineage of cyanopodoviruses isolated from the South China Sea using Prochlorococcus MED4 as the host, and we named this lineage as the MPP-C cluster. The newly isolated MPP-C cyanopodoviruses did not encode DNA pol and auxiliary metabolic genes. They accounted for up to 91% of the cyanopodovirus reads in certain oceanic regions, substantially increasing the total number of hitherto known marine cyanopodoviruses in the global ocean. Furthermore, metagenomic analyses suggested that the MPP-C phages have higher relative abundance in surface ocean than in deeper ocean, which could be linked to their distinct genomic contents and infection kinetics. Our discovery of the overlooked cyanopodovirus group sheds light on the diversity of T7-like viruses in the world’s oceans.
Results
Isolation and genome sequencing of cyanopodoviruses without DNA pol
Using Prochlorococcus MED4 as the host, a high-light-adapted strain abundant in the oceans [49], we isolated 18 cyanopodoviruses from the surface water of the South China Sea, the largest marginal sea with Prochlorococcus as the dominant primary producer [50]. Of the 18 cyanopodoviruses, 17 isolates were highly similar, sharing 95.99% to 99.95% average nucleotide identity (ANI) between each other (Supplementary Figure 1). Since phages with genomic identity above 95% can be considered as the same population [8,9,10], these 17 isolates were grouped into the P-SCSP1 subgroup, with the first P standing for Prochlorococcus, SCS for South China Sea, and the second P for podovirus. The other isolate, P-SCSP2, showed 88.3%–88.4% ANI with the P-SCSP1 subgroup (Supplementary Figure 1, Supplementary Table 1).
The genomes of these newly isolated cyanopodoviruses exhibited a well-conserved T7-like genomic architecture (Fig. 1A), in which three classes of genes were grouped from the left to the right and were shown to be transcribed sequentially in cyanopodoviruses P-SSP7 and S-SBP1 [42, 51]. It is thought that class I genes can modify the host transcriptional machinery, class II genes are mainly involved in DNA metabolism and replication, and class III genes are responsible for phage DNA maturation and virion assembly [51]. DNA pol is always located among class II genes in the 21 previously sequenced cyanopodoviruses (Fig. 1A). However, no DNA pol could be identified in the cyanopodoviruses we isolated from the South China Sea (Fig. 1A).
A. Cyanopodovirus genomes. The phylogenetic tree on the left was generated from the amino acid sequences of the concatenated nine core genes shared by the 39 cyanopodoviruses using a maximum likelihood method. A black dot in a branch indicated bootstrap values larger than 80%. The scale bar represented 0.3 fixed mutations per amino acid position. The names of cyanopodoviruses isolated in this study were shown in red, and those of other phages were shown in black. Cyanophages belonging to different clades were shaded by different colors: orange for MPP-C clade, blue for MPP-B, pink for MPP-A, and grey for P-RSP2. In the MPP-C clade, P-SCSP1a was shown as a representative of cyanophages P-SCSP1a to P-SCSP1u, which share highly similar genomic organizations. In each cyanophage genome, different colors indicated genes with different functions: DNA pol in red, other genes related to DNA metabolism in purple, auxiliary metabolic genes (AMGs) in green, RNA pol in light brown, structural genes in light blue, DNA packaging genes in dark blue, integrase gene in pink, lysozyme gene in dark brown, transcriptional regulator genes in light brown, and genes with unknown function in grey. Three classes of genes, which was shown to be sequentially transcribed in cyanophage P-SSP7, were indicated below the P-SSP7 genome. Abbreviation: SSB, single-stranded DNA binding protein. B Auxiliary metabolic genes in cyanopodovirus genomes. Green and white circles indicated the presence and absence of a gene, respectively. Gene abbreviations: psbA, photosystem II D1 protein; hli, high light inducible protein; MazG, pyrophosphatase; rnr, ribonucleotide reductase; talC, transaldolase; thyX, thymidylate synthase.
Cyanopodoviruses without DNA pol form a new cyanophage MPP-C clade
To study the phylogenetic relationships of cyanopodoviruses with and without DNA pol, we analyzed the core genes shared by all marine cyanopodoviruses. The previously available 21 marine cyanopodoviruses (Table 1) contained 349 unique genes (the pan-genome) and 15 core genes (the core-genome) [40]. After adding the 18 cyanopodovirus genomes we sequenced in this study, the pan-genome of the 39 marine cyanopodoviruses increased to 379 genes (Supplementary Figure 2A) and the core-genome decreased to nine genes (Supplementary Figure 2B). Although the rate of increase seemed to slow down as more genomes were added, the gene accumulation curve was still far from saturated, suggesting undiscovered diversity within cyanopodovirus populations. Of the nine core genes, two encoded proteins involved in DNA metabolism and replication (exonuclease and DNA primase/helicase), five encoded phage structural proteins (major capsid protein, capsid assembly protein, portal protein, tail tube proteins A and B), and two encoded key components of DNA packaging machine (terminase large and small subunits, TerL and TerS, respectively) (Supplementary Figure 3).
The phylogenetic tree based on the concatenated nine core genes divided previously studied cyanopodovirus genomes [34] into the MPP-A and MPP-B clades, with cyanophage P-RSP2 as an outlier (Fig. 1A). The 18 newly isolated cyanopodoviruses without DNA pol formed a distinct clade, branching as a sister lineage to MPP-A and MPP-B clades, which we denoted as the MPP-C clade (Fig. 1A). Phylogenetic trees based on each of the nine core genes also supported the separation of the MPP-C clade from the MPP-A and MPP-B clades (Supplementary Figure 3).
The six genes absent from the previously defined core-genome [40] included two related to virion structural proteins (tail fiber and internal core protein), three involved in DNA metabolism and replication (DNA polymerase, ssDNA binding protein, and endonuclease), and one hypothetical gene. It should be noted that phages of the P-SCSP1 subgroup have genes encoding endonuclease (e.g., gene 06 of P-SCSP1a) that are orthologous to endonucleases of previously known cyanopodoviruses. However, no recognizable gene encoding endonuclease was found in P-SCSP2 (Fig. 1A), implying the divergency of gene content within the MPP-C clade. Gene 05 in P-SCSP1a, which is orthologous to gene 09 in P-SCSP2, is predicted to encode ssDNA binding protein since it shows 47.5% of similarity with ssDNA binding protein of Pelagibacter phage HTVC109P (P109_gp13) [52] via HMMER search (Fig. 1, Supplementary Table 1). However, they do not fall into the same orthologous group with ssDNA binding proteins of previously known cyanopodoviruses. Furthermore, we speculate that g20/g21 of P-SCSP1a and g29/g31 of P-SCSP2 may encode internal virion proteins based on their gene sizes and positions in the genomes. However, none of these four genes can be functionally annotated or show significant hits with genes of published phages. In addition, these four genes show no similarity between P-SCSP1a and P-SCSP2. During infection of coliphage T7, the internal core proteins are thought to form a trans-envelope channel that allows the injection of phage DNA into host cells to initiate infection [53]. The absence of these six genes from the core-genome suggests that the MPP-C phages may have unique replication mechanisms that are distinct from the MPP-A and MPP-B phages.
MPP-C phages do not encode auxiliary metabolic genes and have small genomes
Cyanophages often encode host-like metabolic genes, termed auxiliary metabolic genes (AMGs), whose products were hypothesized to increase phage fitness under stress conditions [54,55,56,57]. Genes for photosystem II D1 protein (psbA), high light inducible protein (hli), pyrophosphohydrolase (mazG), ribonucleotide reductase (rnr), transaldolase (talC), and thymidylate synthase (thyX) have been found in cyanopodovirus genomes [40, 54, 58]. Out of these six AMGs, each of the previously studied cyanopodovirus genomes contained two to five (Fig. 1B) [29, 34, 40]. However, no recognizable AMGs were found in the genomes of MPP-C phages (Fig. 1), suggesting that the MPP-C phages have special metabolic mechanisms and evolutionary trajectories that are distinct from other cyanopodovirus clades.
Consistent with the lack of DNA pol and AMGs, cyanopodoviruses in the MPP-C clade have smaller genomes. Cyanophages in the MPP-A and MPP-B clades had genome sizes of 46.2 ± 0.6 kb and 46.1 ± 1.1 kb, respectively, and the outlier phage P-RSP2 had a genome size of 42.3 kb (Table 1). Cyanophages in the MPP-C clade had genome sizes from 37.3 kb to 37.9 kb (37.8 ± 0.1 kb), which were significantly smaller than those of MPP-A, MPP-B, and P-RSP2 phages (p < 0.001, t-test) (Table 1, Fig. 1). A bioinformatic analysis of double-stranded DNA viruses showed that viruses with small genomes tend not to encode their own DNA polymerase, especially for viral genomes smaller than 40 kb [59]. Our study provides further evidence for the possible correlation between DNA polymerase genes and viral genome sizes.
Host range, morphology, and infection kinetics of MPP-C phages
The host ranges of MPP-C phages were tested against three marine Synechococcus strains (WH7803, WH8012, WH8102) and nine Prochlorococcus strains, of which six belong to the high-light-adapted ecotypes (MED4, AS9601, MIT9312, MIT9202, MIT9215, MIT9301) and three belong to the low-light-adapted ecotypes (MIT9313, NATL1A, NATL2A). We found that the MPP-C phages were highly host-specific and could only infect the original host Prochlorococcus MED4, which is consistent with previous studies showing very narrow host ranges for cyanopodoviruses [12, 34, 35, 43].
Three MPP-C phages (P-SCSP1a, P-SCSP1u, and P-SCSP2) were selected for morphological characterization (Fig. 2A). The transmission electron micrographs of the three phage particles all showed icosahedral capsids and short tails (Fig. 2A), resembling the morphology of the podovirus T7. The capsid diameters of the three MPP-C phages were ~54 nm, slightly smaller than those of Syn5 (60 nm, MPP-A group) and P-SSP7 (65.5 nm, MPP-B group) [41, 60, 61].
A. Transmission electron micrographic images of three representative phages belonging to the MPP-C clade. Scale bars represented 25 nm. B. Infection dynamics of MPP-C (top figures) and MPP-B (bottom figures) phages infecting Prochlorococcus MED4. The abundances of phage DNA in extracellular media were determined using qPCR. The average value and standard deviation of three biological replicates were shown.
To compare the infection kinetics of cyanopodoviruses under identical conditions, we used Prochlorococcus MED4 as the host, which can be infected by both the MPP-C and MPP-B phages. Two MPP-B phages P-SSP7 and P-GSP1 showed similar one step infection dynamics with latent periods of 6–8 h, which are consistent with previous studies of these two phages [51, 62, 63]. Following the latent period, the amounts of extracellular phage DNA for P-SSP7 and P-GSP1 increased exponentially until the plateaus were reached at ~16 hours after infection (Fig. 2B). Compared to MPP-B phages, the MPP-C phages P-SCSP1a and P-SCSP1u had longer latent periods of 12 h and 15 h, respectively, and the extracellular phage DNA reached the plateaus at ~22 h after infection (Fig. 2B). However, despite numerous attempts, we were unable to get a stable one step growth curve for P-SCSP2. The experiments yielded no burst or burst at different time points ranging from ten hours to a few days after inoculation (data not shown). The plateau phase, which represents the end of a single infection cycle, was hardly observed. The variations in lysis time of P-SCSP2 may be attributed to the presence of an integrase gene in its genome, which could facilitate transient integration of cyanophage genome into the host genome [64].
The MPP-C clade is highly abundant in the global surface oceans
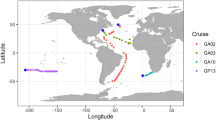
To compare the global distributions of MPP-A, -B, -C, and P-RSP2 phages, we analyzed the viromic datasets of the Tara Oceans project covering different oceanic zones [8, 9]. We also analyzed viromes from surface waters of the South China Sea [65], where the MPP-C phages were isolated. The recruited numbers of cyanopodovirus reads were high in the surface waters of the Indian, Atlantic and Pacific Oceans, and the Mediterranean and Red Seas (Fig. 3A, Supplementary Table 2). Low numbers of cyanopodovirus reads were identified from the Arctic Ocean (Supplementary Table 2), which was reasonable given the low number of cyanobacteria in these regions [66].
A. Relative abundances of the four cyanopodovirus clades in surface waters. Cyanophages in the MPP-A, -B, and -C clades were indicated in pink, blue, and orange, respectively. P-RSP2–like cyanophages were indicated in grey. The size of a pie chart represented the overall recruitment frequency of the cyanopodovirus population in a sampling site, expressed as the total recruited nucleotides of reads (kb) per kb of viral genome per Gb of virome dataset (KPKG). B. The proportions of different cyanopodovirus clades in sampling sites from surface (SRF, 46 sites), deep chlorophyll maximum and the bottom of mixed layer when no deep chlorophyll maximum (DCM + Mix, 33 sites in total). C. Comparison of recruitment frequency of cyanopodovirus clades in the SRF and DCM + Mix sites. Analyses were performed for the 29 sites where both SRF and DCM/Mix waters were sampled. Recruitment frequency was displayed as log10(KPKG + 1). Paired t-tests were applied to compare SRF and DCM + Mix samples: * for p < 0.05, ** for p < 0.01, *** for p < 0.001, and ns for no significance.
In surface waters, the MPP-C phages were highly abundant, accounting for 43.5% of total cyanopodovirus reads across the global ocean, while the contributions of MPP-B and P-RSP2 clades were 52.5% and 3.9%, respectively (Fig. 3B). In the deep chlorophyll maximum (DCM) and the bottom of mixed layer when there was no deep chlorophyll maximum (Mix), the MPP-B clade was dominant (85.3%), followed by MPP-C (11.9%) and P-RSP2 (2.6%) clades (Fig. 3B). In the surface, DCM, and Mix samples, the MPP-A clade was always less than 0.2% of cyanopodovirus reads (Fig. 3B). Globally, the MPP-C phages had higher relative abundance in surface waters than in DCM and Mix waters, while MPP-B phages showed higher relative abundance in DCM and Mix waters than in surface waters (Fig. 3C). P-RSP2–like phages had similar relative abundances in both depths (Fig. 3C).
To further compare the depth profiles of the four cyanopodovirus clades at different months of a year, we analyzed the ALOHA 2.0 viral dataset [67]. In this dataset, a total of 188 virus metagenomes were generated from seawater samples across 1.5 years at 12 depths (5–500 m) in the North Pacific Subtropical Gyre, enabling us to examine the spatiotemporal dynamics of viruses at a high resolution. Consistent with the global patterns (Fig. 3C), the MPP-C phages had higher recruited numbers in waters above 75 m than in deeper waters, while MPP-B phages showed higher recruited numbers between 75 and 100 m, especially during summer and autumn (Fig. 4A). The time-averaged abundances indicated that the recruited numbers of MPP-C phages reached peak at 45 m and MPP-B phages peaked at 75 m (Fig. 4B). The MPP-C phages almost disappeared below 150 m, but MPP-B phages remained detectable between 150 and 250 m (Fig. 4B). The depth profile of MPP-B phages was consistent with a previous PCR-based study reporting that the MPP-B phages in the Red Sea were more abundant at depths [37]. Together, these results indicated that the MPP-C and MPP-B phages have distinct depth profiles in the oceans, with the MPP-C phages showing higher relative abundances in surface waters than in deep water and the MPP-B phages being relatively higher in deeper waters.
Data were recruited from the ALOHA 2.0 viromes covering 0.02–0.2 μm virus-enriched samples across 1.5 years at 12 depths [67]. A Temporal variability in recruitment frequency of different cyanopodovirus clades. Color bar showed the normalized recruitment which was expressed as the total recruited nucleotides of reads (kb) per kb of viral genome per Gb of virome dataset (KPKG). White shading indicated data unavailable from the ALOHA 2.0 viromes. B Depth profiles of month-averaged recruitment frequency of cyanopodoviruses. MPP-A, pink; MPP-B, blue; MPP-C, orange; P-RSP2–like cyanophages in grey.
To assess how environmental variables shaped the global distribution of cyanopodovirus clades, we performed Mantel tests for the viromic datasets with available environmental parameters. The distribution of MPP-C phages was significantly correlated with location (latitude, longitude, depth), temperature, dissolved oxygen, chlorophyll a, nitrite, phosphate concentrations, and Prochlorococcus abundances (Fig. 5A). Among these parameters, the two most positively correlated ones were Prochlorococcus abundances (p < 0.01; r = 0.48) and temperature (p < 0.01; r = 0.45). In addition, the relative importance of environmental variables contributing to the distribution of cyanopodovirus clades was predicted by random forest modeling, which has shown high prediction accuracy in diverse ecological studies [68, 69]. Prochlorococcus abundances showed higher importance in predicting the distribution of MPP-C phages than in predicting other cyanopodovirus clades (Fig. 5B), indicating that the MPP-C phages could be an important predator of Prochlorococcus. Considering the high occurrence of MPP-C phages on a global scale, the tight coupling of MPP-C phages and Prochlorococcus emphasized the potential importance of MPP-C phages in the mortality and community structure of Prochlorococcus in the world’s oceans.
A. The distribution of a cyanopodovirus clade was related to each environmental factor by Mantel’s test. Line colors corresponded to the Mantel’s r statistic for the distance correlations, and line widths denoted the statistical significance based on 999 permutations. Pairwise comparisons of environmental factors with a color gradient denoting Pearson’s correlation coefficient and p values were shown in the circles (*p < 0.05, **p < 0.01, ***p < 0.001). B Variable selection and estimating predictability based on random forest machine-learning algorithm. Histograms represented the percentage of explained variation. Heatmap showed the importance of different variables to the distribution of the four cyanopodovirus clades.
Discussion
In this study, we reported the genomic analyses, infection kinetics, and biogeographic patterns of cyanopodoviruses in the MPP-C clade that we isolated from the South China Sea. The overall genomic architecture of the MPP-C phages resembled that of T7-like cyanopodoviruses in the MPP-A, MPP-B, and P-RSP2 clades. However, the MPP-C phages were the only cyanopodoviruses that do not encode DNA pol or AMGs, and their genomes were the smallest among sequenced cyanophages. Infecting Prochlorococcus MED4, the MPP-C phages showed much longer latent periods than MPP-B phages. By analyzing the global viromic datasets, we revealed that the MPP-C phages contributed to almost half of total cyanopodovirus reads across the global surface ocean. While the MPP-B phages showed higher relative abundances in DCM and Mix waters than in surface waters, the MPP-C phages were relatively more abundant in surface waters than in deep waters. With different depth profiles in the oceans, the MPP-B and MPP-C phages could affect the mortality of Prochlorococcus cells in a broad range of environmental conditions.
The lack of DNA pol suggested that the DNA replication mechanism of the MPP-C phages may be different from that of podoviruses containing DNA pol. The DNA replication complex (replisome) of coliphage T7 is made up of only four proteins: T7 DNA polymerase, T7 single-stranded DNA-binding protein, T7 helicase/primase bifunctional protein, and a processivity factor (thioredoxin) from the host Escherichia coli [70, 71]. While the MPP-A, MPP-B, and P-RSP2 phages all encode their own DNA polymerase and are likely to use similar DNA replication mechanisms as T7, we hypothesize that the MPP-C phages could replicate their genomes by one of three alternative mechanisms. First, the MPP-C phages may recruit the host DNA polymerase I, which belongs to the same DNA polymerase family as those of T7 and cyanopodoviruses. However, DNA polymerase I of prokaryotes is mainly involved in the repair of damaged DNA, but not genome replication [72]. It would be interesting to know whether the MPP-C phages can modify the host DNA polymerase I for viral genome replication. Second, the MPP-C phages may recruit the host DNA polymerase III, which consists of 10 unique subunits and is used by bacteria for genome replication [73]. To recruit host DNA polymerase III for viral DNA replication, Bacillus subtilis phage SPP1 encodes its own initiation protein and several other proteins to interact with the host DNA replication machinery to recognize the viral origin of replication, instead of the host one [74]. The MPP-C phages encode two proteins related to DNA replication (single-stranded DNA-binding protein and DNA helicase/primase bifunctional protein) (Fig. 1A), and thus these proteins may interact with the host DNA polymerase III. Third, we cannot rule out the possibility that the MPP-C phages may encode a DNA polymerase gene that is very divergent from currently known DNA polymerase genes even though no conserved domain corresponding to known DNA polymerase can be detected in the whole genomic sequences of MPP-C phages. Future studies on the DNA replication mechanism of the MPP-C phages will provide novel insights into viral DNA replication processes.
Due to its high conservation in previously sequenced podovirus genomes, DNA pol has been widely used as a genetic marker in quantification and diversity studies of marine cyanopodoviruses [7, 36, 37, 44,45,46]. However, our discovery of the MPP-C phages provided evidence that the T7-like DNA pol cannot fully assess all marine cyanopodoviruses. Thus, we suggest that the nine core genes shared by all marine cyanopodoviruses could serve as suitable genetic markers for marine cyanopodoviruses, especially the major capsid protein (Supplementary Figure 3). The suitable marker genes will help to more accurately measure distinct populations within the marine virioplankton and to quantify virus-mediated ecological processes. For example, in the North Pacific Subtropical Gyre, a polony method using DNA pol estimated that the abundance of cyanopodoviruses was 1.3–4.4 fold lower than that of cyanomyoviruses [45]. Given the high occurrence of MPP-C phages in the global ocean, our finding implies that the abundance of cyanopodoviruses could be much higher than previously thought. In addition, cyanopodovirus-mediated mortality of Prochlorococcus cells in the North Pacific Subtropical Gyre may be higher than estimated using DNA pol [75].
The distribution of viruses in the ocean is generally influenced by the availability of their hosts and the competitive ability of viruses under variable environmental factors [48]. We found that the MPP-C phages were relatively more abundant in surface waters than in deeper waters, while the MPP-B phages were relatively more abundant in deeper layers of the photic zone (Fig. 4B). Similar ecotype-specific depth profiles have been found in Prochlorococcus field populations, with the high-light-adapted ecotypes more abundant in surface waters and the low-light-adapted ecotypes in deeper waters [76]. In our study, the MPP-C phages were isolated from the surface waters using the high-light-adapted ecotype Prochlorococcus MED4 as the host. It remains to be tested whether the abundances of MPP-C and MPP-B phages are correlated to those of different Prochlorococcus ecotypes. Besides the availability of host cells, phages appear to have evolved different strategies to adapt to the environmental availability of resources. For example, the MPP-A, MPP-B, and P-RSP2 cyanopodoviruses encode at least two AMGs which could enable phages to tailor host metabolism and likely provide an advantage for phages to adapt to the environments [54, 77]. However, the MPP-C phages do not encode any of the six auxiliary metabolic genes (Fig. 1B). Two MPP-B phages (P-SSP7 and P-GSP1) isolated on the same host, which both carry AMGs involved in photosynthesis (psbA and hli), nucleotide synthesis (rnr) and carbon metabolism (talC) (Fig. 1), appear to have high relative abundances in both surface waters and deeper waters. While many factors could contribute to this phenomenon, we speculate that light intensity may play a role: host cells in surface waters can derive more energy from light than those in deep waters, thus providing more resources for the propagation of phage progeny, especially for MPP-C phages, which are small and need less resources (e.g., nucleotides). Meanwhile, the existence of AMGs in MPP-B phages is likely to benefit cyanophages at depths with low photosynthetic energy from host cells, enabling the ecological niche expansion of cyanophages in unfavorable conditions.
The biogeographic patterns of phages also depend on their life history traits, such as virion size, host range, infection kinetics, gene content, or lytic/lysogenic infection, etc. [38]. Compared to cyanopodoviruses that we measured in this study and from the literature, MPP-C phages differ in a number of properties, including smaller virions, smaller genome sizes, and longer latent periods. Previous studies suggested that smaller viruses could encounter host cells faster due to greater diffusivity in the marine environment, thus potentially increasing the probability of successful infection [78]. Although it remains to be confirmed whether the sizes of MPP-C phages are smaller than those of all the other cyanopodoviruses, we speculate that the small virion sizes may confer MPP-C phages a competitive advantage in the oligotrophic surface oceans where the abundance of the host is generally low and viral decay is high due to strong UV radiation. In summary, the depth profiles of MPP-C and MPP-B phages may be related to their distinct gene contents and infection kinetics. Similar to Prochlorococcus ecotypes [76], our study suggested that cyanopodoviruses may also evolve into different clades with distinct adaptation mechanisms to enable cyanophages to infect host cells in a broader range of environmental conditions.
Materials and Methods
Phage isolation and purification
The axenic host strain Prochlorococcus MED4 was grown in the Port Shelter (Hong Kong) seawater–based Pro99 [79] medium at 23 °C with continuous light of 25 µmol photons m–2 s–1. The surface seawater samples for phage isolation were collected in December 2015 at the Yongxing Harbor off the Yongxing Island, which is located in the South China Sea (16°50'14.14′)′ N, 112°19'34.30′′ E). The collected seawater samples were filtered through 0.22 μm pore-sized syringe filters and then kept in the dark at 4 °C. To isolate cyanophages infecting Prochlorococcus MED4, filtered seawater samples were mixed with Prochlorococcus MED4 in agarose plates with the helper strain Alteromonas sp. EZ55, following a previously described pour plating method [22]. Plates were incubated at 23 °C under continuous light (25 μmol photons m–2 s–1). Well isolated plaques were picked from the agarose plates, dissolved in the Pro99 medium, and purified with another two rounds of plating to form plaques.
Determining the host ranges of isolated cyanophages
The host ranges of the newly isolated cyanopodoviruses were determined using spot assays [80]. The cyanopodoviruses were challenged against nine Prochlorococcus strains (MED4, MIT9301, MIT9312, MIT9313, AS9601, NATL1A, NATL2A, MIT9215, MIT9202) and three marine Synechococcus spp. strains (WH8012, WH7803, WH8102). Cyanobacterial cultures were plated on agarose plates together with Alteromonas sp. EZ55 as previously described [22]. After the agarose plates solidified, two microliters of each phage lysate were added to the surface of plates. After incubation under continuous light for about one to two weeks, the formation of clear plaques where lysates were added indicates successful phage infection of the cyanobacterial strain in the plate. Tests were repeated at least three times.
Transmission electron microscopy (TEM)
The morphologies of three representative phages (P-SCSP1a, P-SCSP1u and P-SCSP2) were investigated with TEM. In brief, 3 μl of phage lysate were placed on formvar, carbon-coated copper electron microscopy grids and allowed to adsorb for 30 s. Phage particles were negatively stained with 2% (w/v) uranyl acetate for 30 s. Excess stain was removed with a filter paper and grids were air dried before examination with a Thermo-Fisher Talos L120C electron microscope hosted in the Biological Cryo-EM Center of the Hong Kong University of Science and Technology.
Infection kinetics of cyanopodoviruses
Infection kinetics of phages from different cyanopodovirus groups were determined by one step growth experiments [81]. Freshly prepared cyanophage lysates were used to infect exponentially growing Prochlorococcus MED4 cultures (~107 cells per ml) at a multiplicity of infection of 0.1 in triplicate at 21 °C under continuous light (10 µmol photons m–2 s–1). Infected cultures were taken every 2 hours after inoculation and filtered through 0.2 μm polycarbonate filters. Extracellular phage genomic DNA was quantified using quantitative PCR (qPCR), which provides ~1:1 ratio relative to intact phage numbers for both MPP-B and MPP-C phages (Supplementary Table 3). All qPCR assays were run in triplicate following our previous protocol [82]. Each qPCR amplification contained 4.6 μl template, 0.2 μl forward primer (10 μM), 0.2 μl reverse primer (10 μM), and 5 μl iTaq Universal SYBR Green Supermix. Primers were listed in Supplementary Table 4. Reactions were carried out on a LightCycler 480 Real-Time PCR System (Roche). The qPCR program contained one activation step of 95 °C for 5 min followed by 45 amplification cycles of 20 s denaturation at 95 °C and 60 s annealing and elongation step at 60 °C, and a melting curve analysis at the end.
Genome sequencing and gene annotation
To obtain phage DNA for genome sequencing, cyanophage isolates were used to infect Prochlorococcus MED4 cultures to generate 250 ml phage lysates. The phage lysates were filtered through 0.22 μm filters and concentrated using a published FeCl3 flocculation method [83]. After resuspension, the phage concentrates were subjected to dialysis by the TM buffer (0.25 M sucrose, 0.05 M Tris, pH 7.4, 5 mM MgC12). The phage concentrates were treated with 55 μg/ml proteinase K before DNA extraction using the Zymo Clean & Concentrator-5 kit. Viral genomic DNA was sheared into small fragments of around 300 bp in length using the Covaris S220 Focused-ultrasonicator. DNA sequencing libraries were prepared using the NEBNext Ultra II DNA library prep kit and sequenced using the HiSeq 4000 platform (Illumina). The sequencing data generated from each library contains around 2 Gb of 150 bp paired-end reads.
Raw reads were trimmed and filtered using Trimmomatic v0.36 to remove adaptor sequences and low-quality reads [84]. PRINSEQ v0.20.4 was employed to remove redundant sequences [85]. Clean reads were mapped against the Prochlorococcus MED4 genome to remove the host DNA fragments using Bowtie2 [86]. Remaining reads were normalized using bbnorm (https://jgi.doe.gov/data-and-tools/bbtools/) with a target coverage of 150 and assembled using the IDBA assembler v1.1.0 [87] with a maximum k-mer of 130 and a step size of 4. Prokka v1.13 [88] was used to predict and annotate genes in the assembled viral genomes with an e-value of 10-5 and coverage of 50% as the annotation threshold. The complete genome sequences of our newly isolated phages were submitted to the GenBank database (Table 1).
Pairwise average nucleotide identity (ANI) of cyanopodovirus genomes was performed using the pyani (v0.0.12) in ANIm mode (https://pypi.org/project/pyani/). Heatmap was plotted by R (v4.1.2). Core genes of cyanopodoviruses were identified by searching for the protein orthologs shared among the 18 newly sequenced cyanopodovirus isolates and the 21 known cyanopodoviruses using BLASTp and HMM search following previous methods [31]. Briefly, genes were considered orthologs if they were reciprocal best BLASTp hits (e-value ≤ 1e-5) and the alignment covered at least 75% of the length of the shorter gene. The HMM search was further used to identify divergent orthologues that may be missed by the BLAST-based search. The pan- and core-genome curves of the 39 cyanopodoviruses were generated using a random sampling algorithm and plotted as a function of the number of genomes analysed by R. The amino acid sequences of concatenated nine core genes and each individual core gene were used to construct phylogenetic trees to analyze the evolutionary relationships of cyanopodoviruses. Protein sequences were aligned by MUSCLE (v3.8.1551) [89], and phylogenetic trees were constructed with the maximum likelihood method using IQ-TREE (v2.0.3) with 1,000 ultrafast bootstraps [90, 91].
Metagenomics analyses
To estimate the relative abundances and overall distributions of different marine cyanopodovirus clades, fragment recruitment analyses using marine virome data sets from the Tara Oceans expedition, the South China Sea, and the ALOHA station were carried out by BLAST with an e-value <10-5 as described previously [92, 93]. Previous studies have revealed that 95% ANI was a suitable threshold for recovering a set of closely related phage genomes [8, 10, 67]. Hence, in our study, 95% identity was further used to target reads identical to different cyanopodovirus clades across the global oceans. A coverage breadth >90% was set to recruit the mapped reads. If a read was recruited to more than one phage genome, the read was assigned to the phage with the highest bitscore. Recruitment data were normalized according to viral genome size and dataset size to estimate the kb recruited per kb of genome per Gb of metagenome (KPKG) as in previous studies [92, 93]. Phages with <40 % of covered genomes were regarded as absent and given a KPKG value of 0 [94, 95]. Samples with the highest KPKG value less than 0.001 for four clades were considered to have low number of cyanophages and omitted from our subsequent analyses to avoid bias. The global distributions and depth profiles of different cyanopodovirus clades were plotted using the ggplot2 package in R. The Mantel test was performed to assess the correlation between the recruitment frequency of cyanopodoviruses and environmental variables using the R package ‘vegan’. The contributions of different environmental variables in predicting the distribution of cyanopodoviruses were analyzed by random forest analysis using the R package RandomForest.
Data availability
The complete genome sequences of our newly isolated phages are available on the GenBank database under the following accession numbers: OM416763–OM416780. Other Supplementary Information is available for download on the ISME website.
References
Suttle CA. Marine viruses-major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12.
Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature 1999;399:541–8.
Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature 2009;459:207–12.
Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial realm. Nat Microbiol. 2018;3:754–66.
Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, et al. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol. 2020;18:21–34.
Rosenwasser S, Ziv C, Creveld SGV, Vardi A. Virocell metabolism: metabolic innovations during host-virus interactions in the ocean. Trends Microbiol. 2016;24:821–32.
Fuchsman CA, Carlson MCG, Garcia Prieto D, Hays MD, Rocap G. Cyanophage host-derived genes reflect contrasting selective pressures with depth in the oxic and anoxic water column of the Eastern Tropical North Pacific. Environ Microbiol. 2021;23:2782–2800.
Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 2016;537:689–93.
Gregory AC, Zayed AA, Conceição-Neto N, Temperton B, Bolduc B, Alberti A, et al. Marine DNA viral macro-and microdiversity from pole to pole. Cell 2019;177:1109–23.
Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, et al. Patterns and ecological drivers of ocean viral communities. Science 2015;348:1261498.
Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny. Nat Rev Microbiol. 2020;18:125–38.
Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 2003;424:1047–51.
Mann NH. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol Rev. 2003;27:17–34.
Ni T, Zeng Q. Diel infection of cyanobacteria by cyanophages. Front Mar Sci. 2016;2:123.
Flombaum P, Gallegos JL, Gordillo RA, Rincon J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA 2013;110:9824–9.
Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 2015;13:13–27.
Proctor LM, Fuhrman JA. Viral mortality of marine-bacteria and cyanobacteria. Nature 1990;343:60–62.
Carlson MCG, Ribalet F, Maidanik I, Durham BP, Hulata Y, Ferron S, et al. Viruses affect picocyanobacterial abundance and biogeography in the North Pacific Ocean. Nat Microbiol 2022;7:570–80.
Matteson AR, Loar SN, Pickmere S, DeBruyn JM, Ellwood MJ, Boyd PW, et al. Production of viruses during a spring phytoplankton bloom in the South Pacific Ocean near of New Zealand. FEMS Microbiol Ecol 2012;79:709–19.
Ribalet F, Swalwell J, Clayton S, Jimenez V, Sudek S, Lin Y, et al. Light-driven synchrony of Prochlorococcus growth and mortality in the subtropical Pacific gyre. Proc Natl Acad Sci USA. 2015;112:8008–12.
Demory D, Liu R, Chen Y, Zhao F, Coenen AR, Zeng Q, et al. Linking light-dependent life history traits with population dynamics for Prochlorococcus and cyanophage. mSystems 2020;5:e00586–19.
Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 2011;474:604–8.
Marston MF, Pierciey FJ Jr, Shepard A, Gearin G, Qi J, Yandava C, et al. Rapid diversification of coevolving marine Synechococcus and a virus. Proc Natl Acad Sci USA 2012;109:4544–9.
Xiao X, Guo W, Li X, Wang C, Chen X, Lin X, et al. Viral lysis alters the optical properties and biological availability of dissolved organic matter derived from Prochlorococcus picocyanobacteria. Appl Environ Microbiol. 2021;87:e02271–20.
Xiao X, Zeng Q, Zhang R, Jiao N. Prochlorococcus viruses—From biodiversity to biogeochemical cycles. Sci China Earth Sci. 2018;61:1728–36.
Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz JS. The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat Rev Microbiol. 2014;12:519–28.
Puxty RJ, Millard AD, Evans DJ, Scanlan DJ. Viruses inhibit CO2 fixation in the most abundant phototrophs on earth. Curr Biol 2016;26:1585–9.
Weitz JS, Stock CA, Wilhelm SW, Bourouiba L, Coleman ML, Buchan A, et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 2015;9:1352–64.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144.
Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D, et al. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome’. Environ Microbiol. 2009;11:2935–51.
Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol. 2010;12:3035–56.
Sabehi G, Shaulov L, Silver DH, Yanai I, Harel A, Lindell D. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc Natl Acad Sci USA 2012;109:2037–42.
Huang S, Wang K, Jiao N, Chen F. Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage-host genetic exchanges. Environ Microbiol. 2012;14:540–58.
Labrie SJ, Frois-Moniz K, Osburne MS, Kelly L, Roggensack SE, Sullivan MB, et al. Genomes of marine cyanopodoviruses reveal multiple origins of diversity. Environ Microbiol. 2013;15:1356–76.
Dekel-Bird NP, Avrani S, Sabehi G, Pekarsky I, Marston MF, Kirzner S, et al. Diversity and evolutionary relationships of T7-like podoviruses infecting marine cyanobacteria. Environ Microbiol. 2013;15:1476–91.
Huang S, Wilhelm SW, Jiao N, Chen F. Ubiquitous cyanobacterial podoviruses in the global oceans unveiled through viral DNA polymerase gene sequences. ISME J. 2010;4:1243–51.
Baran N, Goldin S, Maidanik I, Lindell D. Quantification of diverse virus populations in the environment using the polony method. Nat Microbiol. 2018;3:62–72.
Chow C-ET, Suttle CA. Biogeography of viruses in the sea. Annu Rev Virol. 2015;2:41–66.
Chen F, Lu JR. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl Environ Microbiol. 2002;68:2589–94.
Huang S, Zhang S, Jiao N, Chen F. Comparative genomic and phylogenomic analyses reveal a conserved core genome shared by estuarine and oceanic cyanopodoviruses. PLoS One. 2015;10:e0142962.
Pope WH, Weigele PR, Chang J, Pedulla ML, Ford ME, Houtz JM, et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: A “horned’ bacteriophage of marine Synechococcus. J Mol Biol. 2007;368:966–81.
Huang S, Sun Y, Zhang S, Long L. Temporal transcriptomes of a marine cyanopodovirus and its Synechococcus host during infection. Microbiologyopen 2021;10:e1150.
Wang K, Chen F. Prevalence of highly host-specific cyanophages in the estuarine environment. Environ Microbiol. 2008;10:300–12.
Chen F, Wang K, Huang S, Cai H, Zhao M, Jiao N, et al. Diverse and dynamic populations of cyanobacterial podoviruses in the Chesapeake Bay unveiled through DNA polymerase gene sequences. Environ Microbiol. 2009;11:2884–92.
Goldin S, Hulata Y, Baran N, Lindell D. Quantification of T4-like and T7-like cyanophages using the polony method show they are significant members of the virioplankton in the North Pacific Subtropical Gyre. Front Microbiol. 2020;11:1210.
Nasko DJ, Chopyk J, Sakowski EG, Ferrell BD, Polson SW, Wommack KE. Family A DNA polymerase phylogeny uncovers diversity and replication gene organization in the virioplankton. Front Microbiol. 2018;9:3053.
Dekel-Bird NP, Sabehi G, Mosevitzky B, Lindell D. Host-dependent differences in abundance, composition and host range of cyanophages from the Red Sea. Environ Microbiol. 2015;17:1286–99.
Hanson CA, Marston MF, Martiny JBH. Biogeographic variation in host range phenotypes and taxonomic composition of marine cyanophage isolates. Front Microbiol. 2016;7:983.
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 2003;424:1042–7.
Chen B, Wang L, Song S, Huang B, Sun J, Liu H. Comparisons of picophytoplankton abundance, size, and fluorescence between summer and winter in northern South China Sea. Cont Shelf Res. 2011;31:1527–40.
Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 2007;449:83–86.
Zhao Y, Qin F, Zhang R, Giovannoni SJ, Zhang Z, Sun J, et al. Pelagiphages in the Podoviridae family integrate into host genomes. Environ Microbiol. 2019;21:1989–2001.
Leptihn S, Gottschalk J, Kuhn A. T7 ejectosome assembly: A story unfolds. Bacteriophage 2016;6:e1128513.
Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA 2011;108:E757–64.
Zeng Q, Chisholm SW. Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr Biol 2012;22:124–8.
Zeng Q, Bonocora RP, Shub DA. A free-standing homing endonuclease targets an intron insertion site in the psbA gene of cyanophages. Curr Biol. 2009;19:218–22.
Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 2005;438:86–89.
Breitbart M, Thompson LR, Suttle CA, Sullivan MB. Exploring the vast diversity of marine viruses. Oceanography. 2007;20:135–9.
Kazlauskas D, Venclovas C. Computational analysis of DNA replicases in double-stranded DNA viruses: relationship with the genome size. Nucleic Acids Res. 2011;39:8291–305.
Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–6.
Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu XG, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 2013;502:707–10.
Liu R, Liu Y, Chen Y, Zhan Y, Zeng Q. Cyanobacterial viruses exhibit diurnal rhythms during infection. Proc Natl Acad Sci USA 2019;116:14077–82.
Maidanik I, Kirzner S, Pekarski I, Arsenieff L, Tahan R, Carlson MCG, et al. Cyanophages from a less virulent clade dominate over their sister clade in global oceans. ISME J. 2022;16:2169–80.
Shitrit D, Hackl T, Laurenceau R, Raho N, Carlson MCG, Sabehi G, et al. Genetic engineering of marine cyanophages reveals integration but not lysogeny in T7-like cyanophages. ISME J. 2022;16:488–99.
Liang Y, Wang L, Wang Z, Zhao J, Yang Q, Wang M, et al. Metagenomic analysis of the diversity of DNA viruses in the surface and deep sea of the South China Sea. Front Microbiol. 2019;10:1951.
Pedrós-Alió C, Potvin M, Lovejoy C. Diversity of planktonic microorganisms in the Arctic Ocean. Prog Oceanogr. 2015;139:233–43.
Luo E, Eppley JM, Romano AE, Mende DR, DeLong EF. Double-stranded DNA virioplankton dynamics and reproductive strategies in the oligotrophic open ocean water column. ISME J. 2020;14:1304–15.
Steidinger BS, Crowther TW, Liang J, Van Nuland ME, Werner GDA, Reich PB, et al. Climatic controls of decomposition drive the global biogeography of forest-tree symbioses. Nature 2019;569:404–8.
Xie X, Wu T, Zhu M, Jiang G, Xu Y, Wang X, et al. Comparison of random forest and multiple linear regression models for estimation of soil extracellular enzyme activities in agricultural reclaimed coastal saline land. Ecol Indic. 2021;120:106925.
Lee SJ, Richardson CC. Choreography of bacteriophage T7 DNA replication. Curr Opin Chem Biol. 2011;15:580–6.
Kulczyk AW, Richardson CC. The replication system of bacteriophage T7. Enzymes. 2016;39:89–136.
Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208.
Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315.
Seco EM, Zinder JC, Manhart CM, Lo Piano A, McHenry CS, Ayora S. Bacteriophage SPP1 DNA replication strategies promote viral and disable host replication in vitro. Nucleic Acids Res. 2013;41:1711–21.
Mruwat N, Carlson MCG, Goldin S, Ribalet F, Kirzner S, Hulata Y, et al. A single-cell polony method reveals low levels of infected Prochlorococcus in oligotrophic waters despite high cyanophage abundances. ISME J. 2021;15:41–54.
Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 1998;393:464–7.
Puxty RJ, Millard AD, Evans DJ, Scanlan DJ. Shedding new light on viral photosynthesis. Photosynth Res. 2015;126:71–97.
Edwards KF, Steward GF, Schvarcz CR. Making sense of virus size and the tradeoffs shaping viral fitness. Ecol Lett. 2021;24:363–73.
Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D, et al. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods. 2007;5:353–62.
Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol. 2010;70:217–48.
Fridman S, Flores-Uribe J, Larom S, Alalouf O, Liran O, Yacoby I, et al. A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat Microbiol. 2017;2:1350–7.
Fang X, Liu Y, Zhao Y, Chen Y, Liu R, Qin QL, et al. Transcriptomic responses of the marine cyanobacterium Prochlorococcus to viral lysis products. Environ Microbiol. 2019;21:2015–28.
John SG, Mendez CB, Deng L, Poulos B, Kauffman AK, Kern S, et al. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ Microbiol Rep. 2011;3:195–202.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20.
Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011;27:863–4.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:1–10.
Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012;28:1420–8.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–9.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic Inference in the genomic era. Mol Biol Evol. 2020;37:2461–2461.
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22.
Martinez-Hernandez F, Fornas O, Lluesma Gomez M, Bolduc B, de la Cruz Pena MJ, Martinez JM, et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat Commun. 2017;8:15892.
Zhang Z, Qin F, Chen F, Chu X, Luo H, Zhang R, et al. Culturing novel and abundant pelagiphages in the ocean. Environ Microbiol 2021;23:1145–61.
Buchholz HH, Michelsen ML, Bolanos LM, Browne E, Allen MJ, Temperton B. Efficient dilution-to-extinction isolation of novel virus-host model systems for fastidious heterotrophic bacteria. ISME J. 2021;15:1585–98.
Qin F, Du S, Zhang Z, Ying H, Wu Y, Zhao G, et al. Newly identified HMO-2011-type phages reveal genomic diversity and biogeographic distributions of this marine viral group. ISME J. 2022;16:1363–75.
Acknowledgements
This study is supported by the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (project number SMSEGL20SC01), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (project number GML2019ZD0409), and the Senior User Project of RV KEXUE (KEXUE2020G09). We acknowledge the support of project SML99147-42080013/99138-42020015 and Center for Ocean Mega-Science, Chinese Academy of Sciences. This study is supported in part by the Project of Hetao Shenzhen-Hong Kong Science and Technology Innovation Cooperation Zone (HZQB-KCZYB-2020083). We thank Changrui Liu and Yuanchao Zhan for assistance in phage isolation and genome sequencing. We thank Prof. Nianzhi Jiao for insightful discussions.
Author information
Authors and Affiliations
Contributions
QZ and LC designed the project; QZ performed seawater sampling and fieldwork with assistance from MH and RZ; LC performed experiments and data analyses; YC sequenced phage genomes and did the pangenome analysis; SX was involved in TEM observation and infection kinetic measurement; LC and QZ wrote the manuscript with contributions from RZ and YC. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cai, L., Chen, Y., Xiao, S. et al. Abundant and cosmopolitan lineage of cyanopodoviruses lacking a DNA polymerase gene. ISME J 17, 252–262 (2023). https://doi.org/10.1038/s41396-022-01340-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-022-01340-6
This article is cited by
-
Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses
Nature Communications (2023)