Abstract
Predatory bacteria are taxonomically disparate, exhibit diverse predatory strategies and are widely distributed in varied environments. To date, their predatory phenotypes cannot be discerned in genome sequence data thereby limiting our understanding of bacterial predation, and of its impact in nature. Here, we define the ‘predatome,’ that is, sets of protein families that reflect the phenotypes of predatory bacteria. The proteomes of all sequenced 11 predatory bacteria, including two de novo sequenced genomes, and 19 non-predatory bacteria from across the phylogenetic and ecological landscapes were compared. Protein families discriminating between the two groups were identified and quantified, demonstrating that differences in the proteomes of predatory and non-predatory bacteria are large and significant. This analysis allows predictions to be made, as we show by confirming from genome data an over-looked bacterial predator. The predatome exhibits deficiencies in riboflavin and amino acids biosynthesis, suggesting that predators obtain them from their prey. In contrast, these genomes are highly enriched in adhesins, proteases and particular metabolic proteins, used for binding to, processing and consuming prey, respectively. Strikingly, predators and non-predators differ in isoprenoid biosynthesis: predators use the mevalonate pathway, whereas non-predators, like almost all bacteria, use the DOXP pathway. By defining predatory signatures in bacterial genomes, the predatory potential they encode can be uncovered, filling an essential gap for measuring bacterial predation in nature. Moreover, we suggest that full-genome proteomic comparisons are applicable to other ecological interactions between microbes, and provide a convenient and rational tool for the functional classification of bacteria.
Similar content being viewed by others
Introduction
Predation is a major evolutionary and ecological force, affecting individual organisms, community structure and whole ecosystems. Our knowledge on the roles and effects of predation mainly relies on numerous studies performed in macro-organisms. In contrast, predation is much less understood in the microbial world, which actually comprises most of the biomass and biological diversity on Earth (Whitman et al., 1998). Nevertheless, some important advances have been achieved: it is now accepted that viral destruction and protozoan grazing are responsible for a large fraction of the microbial turnover in the environment (Fuhrman and Noble, 1995; Pernthaler, 2005); predatory bacteria—bacteria able to feed on other, live bacterial cells—have been shown to respond to changes in prey availability (Chauhan et al., 2009; Chen et al., 2011). Yet, our understanding of the impact of prokaryotic predation on bacterial processes, mortality and dynamics is greatly impaired by our inability to identify novel predatory bacteria, let alone quantify their activity in nature. This stems from several reasons: first, predatory behavior has to be observed by designing predation tests with isolated bacteria, a demanding task that reduces the number of observations. Second, most bacteria are not readily culturable, and this may be even more true for predatory bacteria, which may be unable to grow in the absence of the right prey. Since most prey bacteria are not readily cultured, our ability to grow their predators and observe interactions is greatly compromised. Third, in contrast to other important ecological functions like nitrogen-fixation or photosynthesis, no molecular signatures specific to bacterial predation are known. Thus, the large and expanding metagenomic data obtained from the environment cannot be used to identify putative novel predatory prokaryotes.
So far, the features that have been explored in predatory bacteria and that were shown to be ‘predatory factors,’ that is, functions that affect predatory efficiency (from nullifying it to partially reducing it) were not specific to bacterial predators. They include type IV pili, flagella, chemotactic responses and lytic enzymes (LaMarre et al., 1977; Thomashow and Rittenberg, 1978; Lambert et al., 2003, 2006; Rendulic et al., 2004; Evans et al., 2007) that are also widely found in the genomes of pathogens, saprophytes, autotrophs and others. Although it is possible that specific predatory functions are encoded among the unknown/hypothetical complement genes in the genomes of bacterial predators, another, but not mutually exclusive possibility is that predators’ genomes reflect the predatory phenotype in the distribution and abundance of known genes. We hypothesized that the genomes of prokaryotic predators may be discernable from those of non-predators by a distinctive distribution of known and unknown protein families, and thus provide a mean to detect predators from genome data. To investigate this claim, we surveyed the proteomes of 11 predatory bacteria from across the phylogenetic and ecological landscape against those of 19 non-predators from the same and additional phylogenetic classes. The predators included nine sequenced predatory bacteria and two novel, de novo sequenced genomes, and belonged to the α- and the δ-proteobacteria, Chloroflexi and Bacteroidetes taxa, representing obligate (bacteria unable to grow and complete their life cycle in the absence of prey) and facultative predators, periplasmic (bacteria penetrating and growing in-between the outer and inner membrane of their Gram-negative prey), epibiotic (predators that remain attached to their prey but do not penetrate them) and wolf-pack strategists that lyse prey cells from the outside by concerted action (Martin, 2002).
Materials and methods
Genomes analyzed in this study
The genomes of 11 obligate and facultative predatory bacteria, representing all presently known sequenced genomes of predatory bacteria, and of 19 non-predators originating from the same as well as from different phylogenetic classes were used in this work (Table 1). Non-predators were designated as such based on a literature search. When choosing non-predatory genomes, we had two goals in mind: first, to cover a diverse range of taxa such as the actinobacteria, acidobacteria, firmicutes and chlamydiae; and second, to especially emphasize the proteobacteria, because most of the predators belong to that phylum. The predators include Bdellovibrio and like organisms (BALOs), a group of obligate predatory bacteria that prey on Gram-negative prey, that are epiobiotic or periplasmic (Jurkevitch, 2007). To complete the analysis, two epibiotic BALO species were de novo sequenced by us as described henceforth. Attack phase cells of Bdellovibrio exovorus JSS and Micavibrio aeruginosavorus EPB were obtained from standard lytic cultures prepared using Caulobacter crescentus and Pseudomonas corrugata as prey, respectively (Jurkevitch, 2006). Attack cells were twice filtered through 0.45 μm filters (Sartorius, Goettingen, Germany) to eliminate remnants of the prey populations, and concentrated by centrifugation. Aliquots were spread on NB or PYE plates, and incubated at 30 °C for 3 days to check for any contaminant by observing prey colony formation. Predator DNA was isolated from these cultures with a commercial kit (Promega, Fitchburg, WI, USA) and used for whole-genome paired-ends sequencing with the Genome Analyzer IIx machine (Illumina, San-Diego, CA, USA) at the genome high-throughput sequencing laboratory at Tel Aviv University. Both genomes were assembled by sequentially applying the Abyss (Simpson et al., 2009) and Minimus (Sommer et al., 2007) DNA sequence assemblers. The few resulting contigs were ordered and joined into a single chromosomal sequence by identifying genes and repeats present on the ends of the contigs. The resulting genomes were further analyzed, and corrected when needed, by using the reads pairing data. Directed PCR reactions were used to confirm uncertain short regions and to order the repeats in the B. exovorus JSS CRISPR region. B. exovorus JSS and M. aeruginosavorus EPB underwent open reading frame (ORF) prediction using Prodigal (Hyatt et al., 2010), sequence similarity searches using BLAST (Altschul et al., 1997) and protein domain searches using HMMPFAM (Eddy, 1998). Metabolic reconstruction was performed with Asgard (Alves and Buck, 2007), KAAS (Moriya et al., 2007) and extensive manual curation using BLAST.
Genomic analysis
For each of the analyzed genomes, each putative protein in the genome was classified via BLAST into one of over 120 000 known ortholog protein groups available at the OrthoMCL database (Chen et al., 2006). The OrthoMCL database classifies proteins based on 150 representative complete genomes into orthologous groups; therefore, it is possible for a protein to be ‘unknown’ to OrthoMCL but still produce meaningful BLAST results when searching more comprehensive databases, for example, NCBI. Ortholog groups were created for each species by comparing the species proteomes with the OrthoMCL database, quantifying the number of members each group has for each species. The ortholog groups for the 30 predatory and non-predatory species were arranged in a data matrix where each row was a single species and each column a specific ortholog protein group; each data point in the matrix represented the abundance of the particular ortholog group in the particular genome, relativized to the size of the proteome by dividing in the total number of proteins in that proteome. In total, 12 246 known protein groups were found, averaging 1837±882 groups per proteome. A similar procedure was applied to the proteases, a group of enzymes of special interest in the context of bacterial predation. In each genome, each putative protease was classified into one of 3895 known protease groups within the MEROPS database (Rawlings et al., 2010). A protease group abundance matrix was created for the 30 species as above. The two matrices were separately compared using multivariate analysis in PC-ORD 5.32 software (MjM Software, Gleneden Beach, OR, USA) with Sorensen distances. For all family groups and the proteases our database of microbial species was repeatedly divided into two sets, each time according to a different taxonomic or phenotypic parameter (for example, Bacteria vs Archaea, Gram+ vs Gram−, predatory vs non-predatory bacteria, etc.). After each division, we measured the multivariate difference between the family groups from the two sets; in effect, we performed a multiresponse permutation test (MRPP; Mielke, 1984), which is based on the assumption that, in case the two sets are different from each other, the average within-set difference is smaller than the average between-set distance. The size of the difference between sets was represented by the A-statistic of the MRPP test, while its significance was identified by the MRPP’s P-value. Additionally, cluster analyses were performed with Sorensen distances and flexible β linkages (β=−0.25), and ordinations were performed with non-metric multidimensional scaling (NMDS; Mather, 1976) at 500 iterations, again with Sorensen distances. In order to detect which variables (for example, proteins) were mainly responsible for differences between sets (for example, predators and non-predators) we used the method of Dufrêne and Legendre (1997). The basis for this procedure is the computation of indicator values (IVs) that are a combination of the frequency of occurrence and of the abundance of each variable in each set; IV spans between 0–100, and is larger if a variable is more frequent and/or more abundant in a given set compared with the other set. Usually, as the amount of higher-IV ortholog groups that are included in the test declines, a peak of discriminatory power is reached, after which a drop occurs due to the low number of participating groups. Therefore, multiple MRPP tests using different subsets of ortholog groups (according to their IV) were performed to find a compromise giving the highest possible discrimination power while including as many proteins as possible. Specific proteins in each of the ortholog group in predators and non-predators were manually inspected using BLAST, PFAM (Punta et al., 2009), KEGG pathways (Kanehisa et al., 2011) and ExplorEnz (McDonald et al., 2009). Validation for our system came through using the integrated microbial genomes (IMG) database (Markowitz et al., 2011), which contains full genomes and enables comparison of abundances of specific proteins from different genomes. We chose to compare the abundances of the entire mevalonate and non-mevalonate pathways, containing six enzymes each, between all the 717 finished proteobacterial genomes (representing 409 species) available at IMG. In several instances, manual curation with KEGG pathways followed the IMG automatic annotation. Next, having found ‘indicator proteins’ for both sets, we created a naive ‘predatory index,’ describing how ‘predatory’ each species is. In this index, each species received a +1 point for each of the ‘predatory indicator proteins’ that it contained, and a −1 point for each of the ‘non-predatory indicator proteins’ that it contained.
Data access
Full-genome sequences of M. aeruginosavorus EPB and of B. exovorus JSS were deposited at the NCBI under Mae-EPB.sqn Mae-EPB CP003538 and Bdellovibrio_-JSS.v2e.genome.sqn Bex-JSS CP003537, respectively.
Links to the data are available at http://bioinfo.weizmann.ac.il/~pietro/Bex-JSS.gbf http://bioinfo.weizmann.ac.il/~pietro/Mae-EPB.gbf.
Results
The predators analyzed in this study (Table 1) include BALOs, a group of obligate predatory bacteria that prey on Gram-negative prey, and are epiobiotic or periplasmic (Jurkevitch, 2007). Since available genome sequences were only of periplasmic BALOs, two epibiotic BALO species, B. exovorus JSS, a δ-proteobacterium, and M. aeruginosavorus EPB, a α-proteobacterium, were de novo sequenced to enable a more accurate and complete analysis.
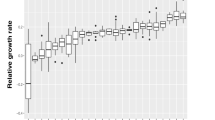
Sequencing of the B. exovorus JSS and M. aeruginosavorus EPB genomes yielded a total number of 37.16 and 34.01 million reads, respectively, with an average length of 36 bp (paired end). Circular chromosomes were assembled, containing 2 657 893 bp with a G+C content of 41.92%, and 2 458 610 bp with a G+C content of 54.96%, for M. aeruginosavorus EPB and B. exovorus JSS, respectively. No extrachromosomal elements were detected. The full exploration of B. exovorus JSS and M. aeruginovorus EPB genomes is beyond the scope of the present study, the focus of which is proteomic abundance profiles. Sequencing of epibiotic BALO predator genomes enabled us to perform a comparison between predators and non-predators with minimal phylogenetic and ecological biases (Table 1). Our analysis shows that there are great differences between predatory and non-predatory proteomes (Figure 1; Supplementary Figure 1); to put these differences in scale, they are much larger than the differences between proteomes of Gram-negative and Gram-positive bacteria, or between those of aerobic and anaerobic bacteria. The differences between proteomes of predators and non-parasitic non-predators are significant, and about half as large as the differences between proteomes of Bacteria and Archaea; incidentally, the degraded proteomes of parasitic intracellular bacteria are extremely different from the proteomes of non-parasites (either predatory or non-predatory). To enable proper evaluation of the results, a phylogeny tree based on the 16S rRNA gene was prepared using the methods described elsewhere (Koval et al., 2012), showing the relatedness of all 30 genomes (Figure 2). In order to detect which protein groups contribute the most to the differences between the proteomes of predatory and of non-predatory species, we employed the ‘IVs’ method of Dufrene and Legendre (1997). This method identified protein families that predominantly appear in the predators set and those that predominantly appear in the non-predators set. IV spans between 0–100, with IV=0 meaning the protein group is equally abundant in both sets whereas IV=100 means that the group is highly abundant in genomes from one set but very rare in genomes from the other set. To increase robustness, the analysis preferably includes as many orthologous groups as possible; however, including more groups inevitably decreases the average IV and reduces the statistical significance of the difference between the predatory and non-predatory sets. To resolve this problem, an optimal compromise was found to be N=31 proteins and minimal IV=69 (Supplementary Figure 2), that is, all the 31 participating protein groups had an IV between 69 and 100, yielding a maximized difference between predatory and non-predatory proteome sets (MRPP A=0.18, P≪0.01). The 31 most indicative proteins, 15 predatory-specific and 16 non-predatory-specific, are listed in Table 2.
Ortholog-based differences between groups of organisms. Differences between protein families within the Bacteria (the six right-most columns) were calculated as the A-statistic of the MRPP test, using 7071 ortholog groups from 27 species. Differences in protein families between Archaea and Bacteria (left-most column) were calculated using 7124 ortholog groups from 30 species (27 bacterial+3 archaeal). *P<0.05.
Phylogenetic tree of the rRNA 16S gene of the 30 genomes analyzed in this study. Gene sequences were retrieved from each genome, aligned by MUSCLE and the evolutionary history was inferred by using the maximum likelihood method in MEGA5. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Predatory bacteria are marked with a gray background.
These 31 ortholog protein groups ( that is, those with IV>69) were used to perform a two-way cluster analysis (Figure 3), after excluding the three archaeal species due to the extreme variation of their genomes from the bacterial ones, and to construct a ‘predatory index’ (Table 3). The cluster analysis, as well as the index, clearly separate the predators from the non-predators without being affected by phylogenetic relatedness, placing all obligate predators, except Micavibrio, at the top of the index list, and confirming the predatory placement of Sorangium, that was originally included as a non-predator in the data set (see Discussion). Note that Acidobacterium was wrongly placed by the clustering algorithm in the predators’ cluster, solely due to the very high abundance of a single uncharacterized protein with a von-Willebrand factor in its genome (Figure 3).
Gene abundance in predators and non-predators. Two-way cluster analysis of genomic abundance of genes encoding for orthologous protein groups which were specific to either predators or non-predators. Complete species and protein data are available in Tables 1 and 2, respectively. Dendrograms were prepared using Sorensen distances and flexible β linkage (β=−0.25).
The predatory-specific protein families may be divided into several broad categories: (1) three mevalonate isoprenoid synthesis pathway enzymes; (2) six adhesion and signaling proteins, including three putative adhesion proteins, potentially involved in cell adhesion and aggregation; two O-linked GlcNAc transferases, proteins known to post-translationally modify signaling proteins in eukaryotes and to control flagellar functions in prokaryotes (Dennis et al., 2006); and a histidine kinase sensor protein; (3) two degradation proteins, including a protease and a benzoate hydrolase; and (4) four metabolism proteins, which may have evolved to scavenge essential metabolites—tryptophan, pyrimidines, flavin and glycerophospholipids. The non-predatory-specific proteins are divided into two categories: (1) five non-mevalonate isoprenoid synthesis enzymes and (2) 11 biosynthesis proteins, involved in biosynthesis of riboflavin (vitamin B2) and of various amino acids. Another difference pointed out by the proteome analysis is that the predators, in contrast to the non-predators, including symbionts, make preferential use of the α2 dimer tRNA glycyl synthetase (GlyRS) and not of the α2/β2 tetramer.
The most striking proteomic difference between predators and non-predators is in their method of synthesizing isoprenoids: all predators except M. aeruginosavorus encode genes coding for the mevalonate pathway but lack genes coding for the non-mevalonate (also known as the 1-deoxy-d-xylulose-5-phosphate or DOXP) pathway, whereas the opposite is true for the non-predators. This finding was independently corroborated by a full proteomic analysis of both isoprenoid pathways: first, all 717 finished proteobacterial genomes (representing 409 species) in the IMG database were queried for the abundance of all six mevalonate pathway enzymes. Out of these 409 sequenced proteobacterial species, 293 (71.6%) have none of the six enzymes, 97 species (23.7%) have one enzyme and 19 species (4.6%) have two or more enzymes. Of the latter, seven have the full DOXP pathway. The remaining 12 ‘mevalonate pathway’ species include all five predatory proteobacterial species available at IMG, plus the intracellular Coxiella burnetii (Omsland et al., 2009), Teredinibacter turnerae (Yang et al., 2009), Cand. Liberibacter asiaticus (Hartung et al., 2011) and Legionella pneumophila (which is also able to grow on dead bacterial cells; Temmerman et al., 2006) (Figure 4). Second, all 717 completed proteobacterial genomes were queried for the abundance of all six non-mevalonate (DOXP) pathway enzymes. Out of the 409 sequenced proteobacterial species, 329 (80.4%) have all six enzymes, and 80 species (19.6%) have one enzyme or none. These 80 ‘DOXPless’ species include all the proteobacterial species found to contain the mevalonate pathway, plus mostly symbionts of arthropods and intracellular pathogens and parasites (such as Rickettsia and Buchnera), which seemed to have altogether lost the ability to synthesize isoprenoids (Supplementary Table 1). The origin of these genes, as seen from a diphosphomevalonoate decarboxylase phylogeny (Supplementary Figure 3), is very different from that of the 16S rRNA gene (Supplementary Figure 4): for example, the λ-proteobacteria predators Sorangium and Bacteriovorax are each closest to different members of the phylum Flavobacteria and Bacteroidetes, while other λ-proteobacteria predators Myxococcus and Stigmatella are closest to members of the phylum Firmicutes, and Bdellovibrio is closest to Legionella. This pattern suggests that the mevalonate pathway was acquired through horizontal gene transfer, by different predator species from different donors. As such, one may hypothesize that the mevalonate pathway confers a real ecological advantage to the predatory lifestyle, and is not merely an evolutionary side-effect.
The isoprenoid pathway in predatory bacteria. The mevalonate pathway compounds (top), enzymes (first row of table, as EC numbers) and enzyme gene abundances in the genomes of 19 proteobacterial species. Known predators are marked in bold. All other 390 finished proteobacterial genomes at the IMG database contained only one or less enzymes of the mevalonate pathway.
In order to further explore the distinction between predators and non-predators and to test the resolution of our analyses, we focused on proteases, specialized proteins known to be present in high numbers in the genomes of predators (Wang et al., 2011; Rendulic et al., 2004; Goldman et al., 2006). The difference in the protease arsenal between predators and non-predators was very large (Supplementary Figure 1B): larger than the full-proteome difference between predators and non-predators, and even larger than the full-proteome difference between Archaea and Bacteria. Indicator analysis, applied to the proteases, showed that the difference between predators and non-predators stems from a small fraction of the families. Thirteen protease families that were very abundant in predators and very rare in non-predators discriminate between these types of bacteria. These families were mainly members of large super families of subtilisins and chymotrypsins, and belonged as well to the aminopeptidase N and peptidoglycan-degrading enzymes (Table 4). When performing the MRPP test between predators and non-predators using only these 13 protease families (instead of the original 123 protease families), the size of the difference doubled to A=0.12.
Discussion
The first complete genome of a predatory bacterium, that of B. bacteriovorus HD100 (Rendulic et al., 2004), provided a glimpse of the haves and haves not of a ‘predatory genome.’ Among the haves, an extended complement of hydrolytic enzymes stood out, along with a large number of transporters, while sensors and regulators were present in average numbers (Rendulic et al., 2004; Tudor and McCann, 2007). On the missing side, biosynthesis pathways for many amino acids as well as for vitamins were absent, and quorum sensing systems were also apparently absent. The analysis of the additional genomes of phylogenetically related and unrelated obligate predators, M. aeruginosavorus strains ARL-13 (GenBank accession number NC_016026) and EPB (this study), B. exovorus JSS (this study) and Bacteriovorax marinus SJ (GenBank accession number FQ312005), confirmed that a lack of biosynthetic capacities to produce many amino acids, as well as vitamins, co-factors and nucleotides to different degrees is a common feature in these obligatorily predatory organisms. Facultative predators also partly lack such biosynthetic capacities (Goldman et al., 2006). Similar characteristics are shared by primary symbionts of higher organisms that lose their ability to produce compounds that can be provided by the host (Moran et al., 2008). Yet, the genomes of obligate symbionts, unlike that of predators, bear the mark of erosion as they are greatly reduced in size, are strongly biased by a low GC content (McCutcheon et al., 2009) and mostly lack DNA repair and recombination genes (Moran et al., 2008). This suggests that a free-living stage (as in facultative symbionts), during which predators are not associated with prey, is linked to preventing genome degradation. Additional instructive missing parts in these genomes include pathogenic and virulence determinants in the forms of effectors, secretion systems (for example, Type III secretion systems) and signaling pathways. Heterotrophic bacteria are able to lyse the constituents of dead cells (Azam and Malfatti, 2007; Martinez et al., 1996) but are not necessarily predatory, thus the question of what underlies the predatory capacity is central to understanding this ability.
Obviously, not all adaptive differences between predatory and non-predatory bacteria are due to difference in gene content. Other source of adaptation, such as differences in gene expression and evolution of ‘core’ genes by point mutation, may play a role in creating and governing the predatory phenomenon. Nonetheless, the current study focuses on comparative analyses of protein orthologs. These are based on relative quantifications and may clearly be biased by the choice and number of the analyzed genomes. Our analysis thus included the genomes of all known sequenced predators along with those of non-predators from the same, as well as from additional, phylogenetic taxa. In order to ascertain whether our chosen 19 non-predator genomes were sufficient to detect real differences between predator and non-predator genomes, we independently corroborated the results using the IMG database with 404 non-predator proteobacterial genomes. Our rationale was that if the same proteins were found to be predator-specific by using both the limited and the extended databases (19 and 404 genomes, respectively), then the results obtained by using the limited database could be trusted. And indeed, the mevalonate/DOXP pathway dichotomy, which was discovered by the limited database, was corroborated by the extended database. This corroboration was pertinent to 25% (8/31) of the proteins shown in Table 2. It is therefore highly likely that if the system proved effective for these proteins, it is effective for all the other ones as well. Similarly, an unavoidable bias of any comparative genomic method is the choice of genomes included, which may affect the results. Thus, one can argue that the ‘predatory index’ described here will only identify novel predatory bacteria that are similar to the 11 already identified. However, the proteome and its predatory index represent the gross genomic investment of an organism in predatory strategy. It shows that while proteomes differ, the commonalities grouping predators together are larger than their differences: Micavibrio, with a proteome deviating in many respects from that of other obligate predators, was still clearly defined as predatory, while Acidobacterium, which weakly clustered with predators (due to high abundances of putative adhesins) was undoubtedly classified as a non-predator by our ‘predatory index’ and analysis of the protease orthologs. Moreover, predators from very different phyla (proteobacteria, Chloroflexi, Bacteroidetes) showed surprising proteomic similarity, while predators and non-predators from the same phylogenetic class (λ-proteobacteria) exhibited proteomic differences as large as the ones between Bacteria and Archaea (Figure 1). The predictive potential of our approach was shown: Sorangium is not mentioned as a predator in the original analysis of its genome (Schneiker et al., 2007), and was thus included in our study as a non-predator. Yet, our analysis clearly revealed its predatory potential. Further search showed that the strong lytic capacities of Sorangium strains against other bacteria were actually demonstrated in 1965 (Gillespie and Cook, 1965). Finally, Saprospira classified high in the predatory index: it appears to be a rather versatile predator able to utilize cyanobacteria or even algal cells (Furusawa et al., 2003; Shi et al., 2006). Although some strains of this organism can grow axenically, many other strains cannot, requiring prey for growth (Sangkhobol and Skerman, 1981) and making them de facto obligate predators.
Bacterial predators exhibit reduced capacities for synthesizing riboflavin and amino acids (mainly tryptophan but also phenylalanine, tyrosine, valine, leucine and isoleucine); biosynthesis proteins for these compounds are strongly underrepresented or totally absent from predators’ genomes, suggesting that they obtain them from the prey. Another significant difference is found in the use of an archeal-eukaryotic GlyRS α2 dimer uncommon in Bacteria (Woese et al., 2000; Wolf et al., 1999), by all obligate and almost all facultative predators instead of the more frequent α2/β2 tetramer composed of two different subunits in non-predators. These two classes lack similarity at the sequence level (Eriani et al., 1990; Wolf et al., 1999). They evolve by different patterns as the tetrameric α and β chains of GlyRS exhibit vertical evolution (Farahi et al., 2004), while the archeal-eukaryotic glyRSs may have been acquired by horizontal gene transfer (Wolf et al., 1999). Alternatively, it may be the ancestral GlyRS that has been displaced by a newly evolved form in the majority of bacteria (Wolf et al., 1999). In addition to depleted protein families such as the metabolic deficiencies mentioned above, the proteomic analysis also discerned protein families enriched in predators in comparison to non-predators. This specialized ‘predatome’ bears a remarkably strong molecular signature apparent in adhesion proteins, lytic enzymes, regulatory factors, and strikingly, in isoprenoid metabolism. The mevalonate pathway for isoprenoid biosynthesis that is found in all higher eukaryotes but few bacteria (Rohmer, 1999) was clearly selected for in almost all predators. It has been suggested that these genes were acquired by B. bacteriovorus by lateral gene transfer (Gophna et al., 2006). Supporting this, a phylogeny of the diphosphomevalonate dicarboxylase shows multiple horizontal acquisitions of the gene by predators. While in the Myxococcales predators (Myxococcus xanthus and Stigmatella auriantica), the gene seems to have a common ancestor close to the Firmicutes, the BALOs Bdellovibrio (Bdellovibrionaceae) and Bacteriovorax (Bacteriovoracaceae), may have acquired it from different organisms. Accordingly, enzymes forming the DOXP pathway, which is used by plants, apicomplexan protozoa and most bacteria, are conspicuously absent from almost all predators and were detected as enriched for in non-predators.
Mevalonate is synthesized from (aceto)acetyl-coA, whereas DOXP uses pyruvate and glyceraldehyde-3-phosphate (Rohmer, 1999). Since the formation of acetyl-coA requires energy that is provided by pyruvate decarboxylation, the former pathway may be advantageous if prey-derived (aceto)acetyl-coA is available. It is noteworthy that almost all free-living non-predators, that may be potential prey, but not the obligate symbionts which are not, possess acetyl-CoA acetyltransferases and are thus a potential source of (aceto)acetyl-coA. Interestingly, hypothetical proteins containing a lipid/polyisoprenoid-binding YceI-like domain, which may be involved in isoprenoid quinone metabolism, transport or storage (Handa et al., 2005) were significantly more abundant in the BALOs, where they are upregulated during growth (Lambert et al., 2010; Wang et al., 2011); also, in myxobacteria a new pathway that branches from 3-hydroxy-3-methylglutaryl-CoA forms isovaleryl coenzyme A and compounds derived thereof, which are essential for fruiting body formation (Lorenzen et al., 2009). These examples of differential enrichment for a metabolic pathway, as well as for a particular tRNA synthetase gene (see above) in phylogenetically unrelated bacterial species suggest that they have been independently selected for and that they may confer as yet unknown selective advantages.
Other enriched functions can be linked to the various needs of predators to bind and degrade prey while regulating their own growth (Table 2). Enriched families of adhesion proteins are strongly enhanced during the free-living attack phase in B. bacteriovorus (Lambert et al., 2010) and have been implicated in adhesion, cell aggregation and heme utilization (Norton et al., 2008). von-Willebrand factor-like (vWF) proteins have been well-studied in eukaryotic organisms, where they are known to be involved in a wide-range of processes including cell-adhesion, transport, the complement system, proteolysis, transcription, DNA repair and ribosome biogenesis. Their roles in Bacteria and Archaea are far less understood, but research has implicated them in bacterial surface adhesion, fibrinogen binding, serum opacity and metal insertion (Kachlany et al., 2000; Katerov et al., 2000; Willows, 2003). Interestingly, preliminary results of proteomic analysis of hypothetical (uncharacterized) proteins suggest that vWF-containing proteins are highly abundant in predator genomes and rarer in non-predator genomes (Pasternak, personal communication).
Prey cell modification and degradation may be brought about by the action of specific proteases that are enriched in predators (Table 4). These enzymes can fulfill complementary functions associated with prey invasion and digestion: M23 zinc metalloproteases are implicated in cell division and in cell reshaping (Bonis et al., 2010; Molle et al., 2010). This family includes endopeptidases of the lysostaphin type that are capable of cleaving Gly–Gly pentaglycin bridges found in the peptidoglycan of Gram-positive bacteria, and DD-endopeptidases that digest D-Ala-diaminopimelic acid cross-linkages (Bonis et al., 2010; Sudiarta et al., 2010) thereby helping in rupturing, and remodeling the prey cell wall. Intramembrane rhomboid proteins are known to cleave near or within trans-membrane domains, and they may be implicated in protein translocation across membranes (Freeman, 2008). Such processes occur in B. bacteriovorus where cell wall proteins are cleaved and a predatory protein is inserted into the prey (Barel et al., 2005). Further modification and breakdown of prey peptidic components into nutrients may be brought about by subtilisins and N-aminopeptidases. In most prokaryotes, secretion of subtilisins outside the cell may provide peptides and amino acids for cell growth or they may help invade host cells (Siezen et al., 2007); the N-aminopeptidases release N-terminal amino-acid residues, breakdown exogenously supplied peptides, and participate in the final steps of protein turnover, enabling the utilization of amino acids as nutrients (Ito et al., 2006; Kumar and Nandi, 2008). Accordingly, in B. bacteriovorus and Micavibrio, genes encoding these enzymes are produced during growth on a prey substrate and may thus be associated with internal degradation of the host (Lambert et al., 2010; Wang et al., 2011), along with the very strongly predator-enriched chymotrypsin proteases (Dori-Bachash et al., 2008; Lambert et al., 2010; Wang et al., 2011). In addition to prey remodeling and consumption, proteases such as S2P proteases, and O-linked GlcNAc transferases can coordinate various regulatory cell growth and differentiation functions (Wolfe and Kopan, 2004), including flagellar gene expression and flagellar assembly in the latter (Shen et al., 2006; VanDyke et al., 2009). In the obligate predators B. bacteriovorus and M. aeruginosavorus different homologs of the former family are expressed during the free swimming and during the attachment phases (Lambert et al., 2010; Wang et al., 2011).
The science of dividing microorganisms into groups has passed through the two major phases of ‘phenotypic taxonomy’ that spanned from the mid-19th century to the 1970s and relied on morphological and chemical traits to define microbes (Schleifer and Kandler 1972), and of ‘limited genomic taxonomy,’ that occurred from the 1970s to the present day, and relies on DNA–DNA hybridization (De Ley et al., 1970) and 16S rDNA cataloging (Fox et al., 1977). The recent advent of high-throughput sequencing has greatly increased the availability of whole-genome sequences (Hall, 2007), drastically changing the way microbiologists study microbes, in what has been termed the ‘-omics revolution’ (MacKenzie, 2001). This revolution, however, seems to have been largely ignored by the field of microbial taxonomy, mainly due to the (i) current lack of sequenced genomes from many major prokaryotic lineages and (ii) significant amount of lateral gene transfer in prokaryotic genomes (Klenk and Göker, 2010). With both sequencing and analysis techniques rapidly improving, microbial taxonomy may be in the beginning of its third phase—‘functional taxonomy’—which would rely on genome-wide comparisons of genes (Kislyuk et al., 2011) and/or proteins (Callister et al., 2008). Our study applies full-genome proteomics in studying the predatory bacteria and identifying core proteins. This approach may then prove useful for classifying known predatory bacteria as well as for finding new ones in nature, two tasks that have thus far proved quite difficult. Consequently, most of our knowledge still originates from a few taxa in the δ-proteobacteria, that is, the BALOs and the myxococcales, the observation and analysis of which suggest that bacterial predators may affect bacterial mortality (Chauhan et al., 2009; Chen et al., 2011), and offer potential treatments against Gram-negative ailments (Atterbury et al., 2011). Yet, without a more realistic assessment of their abundance and diversity, a true understanding of their impact in nature cannot be achieved. Our approach of using full-genome proteomic comparisons offers a novel tool for functionally classifying known predatory bacteria as well as for exploiting (meta)genomic data and uncovering novel predators. We suggest that such functional taxonomical approach may also be largely applicable to other ecological interactions involving microbes.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25: 3389–3402.
Alves JMP, Buck GA . (2007). Automated system for gene annotation and metabolic pathway reconstruction using general sequence databases. Chem Biodiv 4: 2593–2602.
Atterbury RJ, Hobley L, Till R, Lambert C, Capeness MJ, Lerner TR et al (2011). Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol 77: 5794–5803.
Azam F, Malfatti F . (2007). Microbial structuring of marine ecosystems. Nat Rev Microbiol 5: 782–791.
Barel G, Sirota A, Volpin H, Jurkevitch E . (2005). Fate of predator and prey proteins during growth of Bdellovibrio bacteriovorus on Escherichia coli and Pseudomonas syringae prey. J Bacteriol 187: 329–335.
Bonis M, Ecobichon C, Guadagnini S, Prévost M, Boneca IG . (2010). A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol 78: 809–819.
Callister SJ, McCue LA, Turse JE, Monroe ME, Auberry KJ, Smith RD et al (2008). Comparative bacterial proteomics: analysis of the core genome concept. PLoS One 3: e1542.
Chauhan A, Cherrier J, Williams HN . (2009). Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. PNAS 106: 4301–4306.
Chen F . (2006). OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucl Ac Res 34: D363–D368.
Chen H, Athar R, Zheng G, Williams HN . (2011). Prey bacteria shape the community structure of their predators. ISME J 5: 1314–1322.
De Ley J, Cattoir H, Reynaerts A . (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12: 133–142.
Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ et al (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol 13: 365–371.
Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E . (2008). Proteome-based comparative analyses of growth atages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl Environ Microbiol 74: 7152–7162.
Dufrêne M, Legendre P . (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Mono 67: 345–366.
Eddy SR . (1998). Profile hidden Markov models. Bioinformatics 14: 755–763.
Eriani G, Delarue M, Poch O, Gangloff J, Moras D . (1990). Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347: 203–206.
Evans KJ, Lambert C, Sockett RE . (2007). Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol 189: 4850–4859.
Farahi K, Pusch G, Overbeek R, Whitman W . (2004). Detection of lateral gene transfer events in the prokaryotic tRNA synthetases by the ratios of evolutionary distances method. J Mol Evol 58: 615–631.
Fox GE, Magrum LJ, Balch WE, Wolfe RS, Woese CR . (1977). Classification of methanogenic bacteria by 16S ribosomal RNA characterization. PNAS 74: 4537–4541.
Freeman M . (2008). Rhomboid proteases and their biological functions. Ann Rev Gen 42: 191–210.
Fuhrman JA, Noble RT . (1995). Viruses and protists cause similar bacterial mortality in coastal seawater. Limonol Oceanog 40: 1236–1242.
Furusawa G, Yoshikawa T, Yasuda A, Sakata T . (2003). Algicidal activity and gliding motility of Saprospira sp SS98-5. Can J Microbiol 49: 92–100.
Gillespie DC, Cook FD . (1965). Extracellular enzymes from strains of Sorangium. Can J Microbiol 11: 109–118.
Goldman BS . (2006). Evolution of sensory complexity recorded in a Myxobacterial genome. PNAS 103: 15200–15205.
Gophna U, Charlebois RL, Doolittle WF . (2006). Ancient lateral gene transfer in the evolution of Bdellovibrio bacteriovorus. Tr Mcrobiol 14: 64–69.
Hall N . (2007). Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol 210: 1518–1525.
Handa N, Terada T, Doi-Katayama Y, Hirota H, Tame JRH, Park SY et al (2005). Crystal structure of a novel polyisoprenoid binding protein from Thermus thermophilus HB8. Prot Sci 14: 1004–1010.
Hartung JS, Shao J, Kuykendall LD . (2011). Comparison of the candidatus liberibacter asiaticus’ genome adapted for an intracellular lifestyle with other members of the Rhizobiales. PLoS One 6: e23289.
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ . (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119.
Ito K, Nakajima Y, Xu Y, Yamada N, Onohara Y, Ito T et al (2006). Crystal structure and mechanism of tripeptidyl activity of prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis. J Mol Biol 362: 228–240.
Jurkevitch E . (2006). The Genus Bdellovibrio. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, (eds.) The Prokaryotes. Springer: New York, pp 12–30.
Jurkevitch E . (2007). Predatory behaviors in bacteria—diversity and transitions. Microbe Am Soc Microbiol 2: 67–73.
Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH et al (2000). Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J Bacteriol 182: 6169–6176.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M . (2011). KEGG for integration and interpretation of large-scale molecular data sets. Nucl Ac Res 40: D109–D114.
Katerov V, Lindgren P-E, Totolian AA, Schalén C . (2000). Streptococcal opacity factor: a family of bifunctional proteins with lipoproteinase and fibronectin-binding activities. Curr Microbiol 40: 149–156.
Kislyuk AO, Haegeman B, Bergman NH, Weitz JS . (2011). Genomic fluidity: an integrative view of gene diversity within microbial populations. BMC Genomics 12: 32.
Klenk H-P, Göker M . (2010). En route to a genome-based classification of Archaea and Bacteria? System Appl Microbiol 33: 175–182.
Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E . (2012). Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int J Syst Evol Microbiol ijs.0.039701-0.
Kumar A, Nandi D . (2008). Characterization and role of peptidase N from Salmonella enterica serovar Typhimurium. Bioch Biophys Res Comm 353: 706–712.
LaMarre AG, Straley SC, Conti SF . (1977). Chemotaxis toward amino acids by Bdellovibrio Bacteriovorus. J Bacteriol 131: 201–207.
Lambert C, Chang C-Y, Capeness MJ, Sockett RE . (2010). The First Bite—Profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5: e8599.
Lambert C, Evans KJ, Till R, Hobley L, Capeness M, Rendulic S et al (2006). Characterizing the flagellar filament and the role of motility in bacterial prey penetration by Bdellovibrio bacteriovorus. Mol Microbiol 60: 274–286.
Lambert C, Smith MCM, Sockett RE . (2003). A novel assay to monitor predator–prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl accepting chemotaxis proteins in predation. Environ Microbiol 5: 127–132.
Lorenzen W, Ring MW, Schwär G, Bode HB . (2009). Isoprenoids are essential for fruiting body formation in Myxococcus xanthus. J Bacteriol 191: 5849–5853.
MacKenzie S . (2001). High-throughput interpretation of pathways and biology. Drug News Perspect 14: 54–57.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al (2011). IMG: the integrated microbial genomes database and comparative analysis system. Nucl Ac Res 40: D115–D122.
Martin MO . (2002). Predatory prokaryotes: an emerging research opportunity. J Mol Microbiol Biotechnol 4: 467–477.
Martinez J, Smith DC, Steward GF, Azam F . (1996). Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol 10: 223–230.
Mather PM . (1976) Computational Methods of Multivariate Analysis in Physical Geography. J Wiley and Sons: London, England, 532.
McCutcheon JP . (2009). Convergent evolution of metabolic roles in bacterial co-symbionts of insects. PNAS 106: 15394–15399.
McDonald AG, Boyce S, Tipton KF . (2009). ExplorEnz: the primary source of the IUBMB enzyme list. Nucl Ac Res 37: D593–D597.
Mielke PW . (1984) 34 Meteorological Applications of Permutation Techniques Based on Distance Functions, in Nonparametric Methods. Elsevier, 813–830.
Molle V, Leiba J, Zanella-Cléon I, Becchi M, Kremer L . (2010). An improved method to unravel phosphoacceptors in Ser/Thr protein kinase-phosphorylated substrates. Proteomics 10: 3910–3915.
Moran NA, McCutcheon JP, Nakabachi A . (2008). Genomics and evolution of heritable bacterial symbionts. Ann Rev Gen 42: 165–190.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M . (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucl Ac Res 35: W182–W185.
Norton JM, Klotz MG, Stein LY, Arp DJ, Bottomley PJ, Chain PS et al (2008). Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl Environ Microbiol 74: 3559–3572.
Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE et al (2009). Host cell-free growth of the Q fever bacterium Coxiella burnetii. PNAS 106: 4430–4434.
Pernthaler J . (2005). Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3: 537–546.
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J et al (2009). MEROPS: the peptidase database. Nucl Ac Res 38: D227–D233.
Rawlings ND, Barrett AJ, Bateman A . (2010). ‘MEROPS: the peptidase database’. Nu Ac Res 38: D227–D233.
Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C et al (2004). A predator unmasked: Life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303: 689–692.
Rohmer M, Rohmer M . (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Pro Rep 16: 565–574.
Sangkhobol V, Skerman VBD . (1981). Saprospira species—natural predators. Cur Microbiol 5: 169–174.
Schleifer KH, Kandler O . (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36: 407–477.
Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO et al (2007). Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotech 25: 1281–1289.
Shen A, Kamp HD, Gründling A, Higgins DE . (2006). A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Gen Dev 20: 3283–3295.
Shi M, Zou L, Liu X, Gao Y, Zhang Z, Wu W et al (2006). A novel bacterium Saprospira sp strain PdY3 forms bundles and lyses cyanobacteria. Front Biosci 11: 1916–1923.
Siezen RJ, Renckens B, Boekhorst J . (2007). Evolution of prokaryotic subtilases: genome-wide analysis reveals novel subfamilies with different catalytic residues. Prot Struc Func Bioinf 67: 681–694.
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol İ . (2009). ABySS: a parallel assembler for short read sequence data. Gen Res 19: 1117–1123.
Sommer DD, Delcher AL, Salzberg SL, Pop M . (2007). Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8: 64.
Sudiarta IP, Fukushima T, Sekiguchi J . (2010). Bacillus subtilis CwlP of the SP-B prophage has two novel peptidoglycan hydrolase domains, muramidase and cross-linkage digesting dd-endopeptidase. J Biol Chem 285: 41232–41243.
Temmerman R, Vervaeren H, Noseda B, Boon N, Verstraete W . (2006). Necrotrophic growth of legionella pneumophila. Appl Environ Microbiol 72: 4323–4328.
Thomashow MF, Rittenberg SC . (1978). Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: solubilization of Escherichia coli peptidoglycan. J Bacteriol 135: 998–1007.
Tudor J, McCann M . (2007). Genomic analysis and molecular biology of predatory prokaryotes. In Jurkevitch E, (eds.) Predatory Prokaryotes Microbiology Monographs. Springer: Berlin, Heidelberg, pp 153–189.
VanDyke DJ, Wu J, Logan SM, Kelly JF, Mizuno S, Aizawa S et al (2009). Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol Microbiol 72: 633–644.
Wang Z, Kadouri DE, Wu M . (2011). Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics 12: 453.
Whitman WB, Coleman DC, Wiebe WJ . (1998). Prokaryotes: the unseen majority. PNAS 95: 6578–6583.
Willows RD . (2003). Biosynthesis of chlorophylls from protoporphyrin. IX Nat Prod Rep 20: 327–341.
Woese CR, Olsen GJ, Ibba M, Söll D . (2000). Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev 64: 202–236.
Wolf YI, Aravind L, Grishin NV, Koonin EV . (1999). Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Gen Res 9: 689–710.
Wolfe MS, Kopan R . (2004). Intramembrane proteolysis: theme and variations. Science 305: 1119–1123.
Yang JC, Madupu R, Durkin AC, Ekborg NA, Pedamallu CS, Hostetler JB et al (2009). The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (Shipworms). PLoS One 4: e6085.
Acknowledgements
We thank Susan Koval at the University of Western Ontario and Albina Afinogenova (previously at the Russian Academy of Sciences, Pushchino, Russia) for kindly providing B. exovorus JSS and M. aeruginosavorus EPB, respectively. This work was supported by a grant of the Israel Science Foundation (Grant No1290/08).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Pasternak, Z., Pietrokovski, S., Rotem, O. et al. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J 7, 756–769 (2013). https://doi.org/10.1038/ismej.2012.149
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2012.149
Keywords
This article is cited by
-
Taxonomic and environmental distribution of bacterial amino acid auxotrophies
Nature Communications (2023)
-
Predatory bacteria as potential biofilm control and eradication agents in the food industry
Food Science and Biotechnology (2023)
-
High impact of bacterial predation on cyanobacteria in soil biocrusts
Nature Communications (2022)
-
Effects of straw mulching on predatory myxobacterial communities in different soil aggregates under wheat-corn rotation
Environmental Science and Pollution Research (2022)
-
Venatorbacter cucullus gen. nov sp. nov a novel bacterial predator
Scientific Reports (2021)