Abstract
Community similarity declines with increasing geographic distance if species tend to be locally adapted or if they are dispersal limited. The distance–decay of similarity has been shown for bacteria previously, but distinguishing between these competing mechanisms is difficult from observational surveys. I found little evidence of a relationship between geographic distance and similarity in community composition in an aquatic bacterial community. When bacterial colonization occurred into initially identical sterile microcosms across a woodland, a strong distance–decay relationship was observed after 28 days, implying that dispersal limitation created a strong pattern in these communities in the absence of environmental differences. This conclusion was not supported by the results of a reciprocal transplant experiment. When microcosms at the extremities of the study area were transplanted to the opposite end of the study area, the communities converged on the community composition at the site to which they were transplanted. This convergence did not depend on whether colonization into the microcosms was prevented, implying a minor role for dispersal limitation over these spatial and temporal scales. Additional manipulations of colonization rates were consistent with the hypothesis that dispersal limitation structured these communities over short time scales (a few days), but that dispersal limitation had a minor role over longer time scales (>7 days).
Similar content being viewed by others
Introduction
Ecological communities are likely to become increasingly different as the distance between the communities increases (Nekola and White, 1999). In general, two mechanisms might generate this pattern of declining similarity with increasing geographical distance. First, environmental conditions are likely to become increasingly different with distance. If the species within a community are adapted to local conditions, communities are expected to become increasingly different as species are sorted according to their niche requirements. Under this scenario, dissimilarity among communities simply parallels the increasing disparities in current (or recent) environmental conditions with increasing geographical distance. Second, individuals are dispersal limited. If individuals tend to colonize nearby sites, sites that are close together will tend to have similar communities even if the species are ecologically equivalent. In this scenario, a distance–decay relationship would emerge even without differences in environmental conditions or niche requirements (Bell, 2001; Hubbell, 2001).
Attempts to describe the distance–decay relationship and to disentangle the ecological mechanisms involved have relied on comparing observed patterns of distance–decay with model expectations (Nekola and White, 1999; Bell, 2001; Morlon et al., 2008). For example, one approach is to analyze whether there is a relationship between community similarity and distance once variation because of known environmental factors has been removed (Legendre et al., 2005). Unfortunately, identifying the environmental factors that determine the distribution of abundance of every species in a community is arduous, especially for species-rich communities in which the ecology of the constituent species is poorly understood. Observational studies provide an important starting point for generating hypotheses regarding the relative importance of dispersal limitation versus local adaptation, but are unlikely to identify causal mechanisms without accompanying experimental manipulations. Conversely, although manipulative experiments would allow stronger inferences, such experiments are not possible for most ecological communities.
Microbial communities offer an excellent opportunity to test the ecological mechanisms that underlie macroecological patterns because both observational studies and manipulative experiments are possible under natural conditions (Jessup et al., 2004, 2005). In the past decade, there have been a number of studies on the biogeography of microbial species, many of which have indicated some degree of endemicity (Green and Bohannan, 2006; Martiny et al., 2006). Thus, although it may be that many microbial species are globally distributed, most surveys that use molecular techniques to identify microbial taxa have shown that many microbial species are also limited in spatial extent in the same way as larger organisms (Martiny et al., 2006; Pommier et al., 2007). Studies have now identified spatial patterns at spatial scales that range from a few centimetres (Franklin and Mills, 2003) to hundreds of kilometres (Cho and Tiedje, 2000; Franklin and Mills, 2003; Pommier et al., 2007). There have now been several observational studies that attempt to distinguish between environmental versus dispersal limitation of bacterial communities (Green et al., 2004; Horner-Devine et al., 2004; Legendre et al., 2005; Ramette and Tiedje, 2007; Van der Gucht et al., 2007; Sommaruga and Casamayor, 2009), and have generally found signals of significant environmental effects on species spatial turnover, whereas evidence of dispersal limitation has been implicated particularly at large spatial scales (Whitaker et al., 2003; Green et al., 2004). Increasingly, there have been efforts to describe environmental associations of bacterial taxa to piece together the natural history of broad taxonomic groups (Fierer et al., 2007; Suen et al., 2007; Hunt et al., 2008), but such studies remain rudimentary because of the enormous taxonomic richness of these communities. In addition, even if the natural history was well described, there is still the problem that variation in the environment is likely to be partially confounded with geographic distance, making it difficult to distinguish between these two factors. This study adds to this growing literature by combining an investigation of spatial patterns in naturally occurring bacterial communities with experimental manipulations to test the mechanisms involved.
Microcosm experiments using artificially assembled communities have often been regarded as too contrived to have much applicability to ‘real-world’ ecological communities (Carpenter, 1996). However, it is also possible to conduct experimental manipulations in situ using natural communities of microbes, an approach that does not suffer from this criticism and which offers a convenient intermediate between artificially constructed microcosm communities and the full complexity of natural communities. This study focuses on the bacterial communities that inhabit pools of rainwater formed at the base of beech trees in southern England. These are often permanent bodies of water with a volume up to several litres. Previous research has identified a strong relationship between bacterial taxonomic diversity and the volume of these pools of rainwater, and that the form of the relationship is similar to the power law relationship frequently observed for larger organisms (Bell et al., 2005). Further analysis of the same data set showed that a neutral model could explain the distribution of abundance in these communities based solely on differences in the number of individuals in the communities (Woodcock et al., 2007). Importantly, the study showed that the same parameter values could be used to fit models across the tree-hole communities, implying that the same ecological factors were structuring the tree-hole communities within the woodland. These observational findings are consistent with the hypothesis that dispersal limitation and differences in community size were sufficient to explain differences in the composition of these communities.
Under the hypothesis that dispersal limitation explains the spatial distribution of the bacteria in this woodland, I conducted experiments that attempted to test the following predictions: (1) bacterial communities that are increasingly distant will have communities that are increasingly dissimilar; (2) this distance–decay in similarity occurs even in the absence of environmental differences; (3) communities that are transplanted to new locations will take on a taxonomic composition of nearby communities; (4) communities that are transplanted to new locations will not take on a composition similar to nearby communities if colonization is prevented; and (5) artificially modulating the colonization rate will affect the final community composition.
Materials and methods
The tree-hole micro-ecosystem
I used an aquatic micro-ecosystem to test the distance–decay relationship. The buttressing of large European beech trees (Fagus sylvatica) often trap rainwater. The resulting rain-pools are ephemeral in exposed locations, but can be permanent if sufficiently shaded. The beech leaves and mast that fall into the tree-holes provide an energy source that supports an array of organisms, including a specialized larval insect community (Kitching, 1971). The aquatic communities are assumed to represent discrete, aquatic islands embedded within the surrounding terrestrial environment. These natural microcosms (Srivastava et al., 2004) have been used as convenient replicated communities to test ecological theory (Bradshaw and Holzapfel, 1992; Srivastava and Lawton, 1998). The principal mode of bacterial colonization into the tree-holes is unknown, but it seems reasonable to assume that colonization occurs particularly after rainfall, and occasionally through deposition from the air, or by being transported by other organisms.
Distance–decay in water-filled tree-holes
I conducted surveys of the tree-hole communities every 2 days for 30 days from 10 July 2008. The surveys were conducted in tree-holes that encompassed the entirety of the main concentration of beech trees within Wytham Woods, Oxfordshire. Tree-holes that had not dried up over the previous 2 years were selected for the study. This limited the study to relatively large tree-holes and minimized the variation in tree-hole volume, which has previously been shown to influence bacterial diversity (Bell et al., 2005). The tree-hole surface area (which is strongly correlated with volume) did not vary systematically across the study area. During each survey, the contents of each tree-hole was homogenized using a sterile pipette and a 2-ml sample was taken for molecular analysis (see below). Tree-hole locations were measured using Garmin GPSMAP 76CSx, and distance between tree-hole locations was estimated using MapSource version 6 (Garmin, Southamptom, UK).
Distance–decay in microcosms
Observations of natural communities are unlikely to disentangle the ecological mechanisms responsible for explaining spatial patterns because several ecological mechanisms produce the same observational patterns. I therefore conducted a series of microcosm field experiments to identify the mechanisms responsible for distance–decay patterns. Freshly fallen beech leaves were collected during 2006 and kept at −20 °C until the initiation of the experiment. A beech-leaf broth medium was created by autoclaving 0.1 g beech-leaf litter per ml water (Volvic mineral water, Sainsburys Supermarkets Ltd, London, UK). The broth was filtered to remove particulate matter and diluted 16-fold in water. Glass microcosms were amended with 10 ml of the diluted sterile beech-leaf broth and placed in the Wytham Woods in the same stand of beech trees as the survey (above). A total of 112 microcosms were placed 71-m apart (that is, at: 0, 71, 142, 213, 284, 355, 426, 497 m) so that they were distributed throughout the entirety of the study area (14 microcosms per site). A 2-ml sample was taken from each microcosm after 14 and 28 days, which was used for the molecular analysis (below). Of the 14 replicates at each site, 7 were covered with sterile caps after 14 days until the end of the experiment (day 28) to prevent further colonization.
Reciprocal transplant experiment
Simply documenting the spatial pattern across the study area while attempting to control the environmental variables is insufficient to show conclusively that dispersal limitation occurs. Although effort was made to ensure that the environment was constant in microcosms across the study area, I could not exclude the possibility that gradients of light, temperature or other environmental variables were present. I therefore conducted a reciprocal transplant experiment to identify the relative importance of dispersal limitation versus environmental factors in determining the dissimilarity among microcosm communities. Forty additional microcosms were placed at each extremity of the study area. After allowing 14 days of colonization, 20 microcosms from each site were transported to the other site (that is, a reciprocal transplant experiment). Also at the 14-day mark, of the 80 microcosms, half were closed to colonization using a sterile cap. The experiment therefore consisted of a fully factorial design with three treatments (site, colonization and transplanted) each with two levels (site 1 or 2, open or closed to colonization, transplanted or not) each of which was replicated 10 times.
For the microcosms closed to colonization, any change in the communities (relative to those that remain at the original site) must be due to something other than dispersal. Therefore, by definition, any change in the communities must be due to some environmental component. A second possibility is that cutting off colonization would result in a change in the community composition. This would occur if dispersal was important in maintaining the observed community composition. If the observed community composition was solely due to the continual influx of colonizers, the community composition would be expected to change substantially once immigration into the microcosms was eliminated. Finally, it is possible that the amount of change in community composition after transplantation would depend on whether or not the microcosm was open to colonization (that is, whether there was a colonization × transplantation interaction). Such a result would imply that the response of the community following changes to the environment relies on colonization. Reciprocal transplant studies that simultaneously manipulate colonization therefore provide an important tool for estimating the relative importance of dispersal limitation and environmental conditions on community composition.
Manipulation of colonization rate
If environmental conditions alone determined community composition, manipulating the colonization rate into microcosms at a specific location (that is, constant environment) should have no effect on the final composition. I manipulated colonization rates into microcosms by opening and closing the microcosms at different rates. For this experiment, 128 microcosms were placed at the centre of the study area. Microcosms were closed to colonization by loosely capping the microcosms with sterile screw-on caps. Caps were added or removed at 2-day intervals. There were 16 levels of colonization rate (caps open for 0, 2, 4, …, 28 or 30 days) with 8 replicates per colonization level. Within the constraint of the colonization rate treatment assigned to each microcosm, the pattern of cap removal/replacement was determined at random. The sterile zero-colonization controls were uncontaminated at the end of the experiment and so were excluded from further statistical analysis.
Molecular analysis of bacterial communities
Samples were returned to the laboratory on the day they were collected. The samples were centrifuged for 10 min at 13 000 revolutions per minute, the liquid phase was decanted and the samples were then stored at −20 °C until the conclusion of the experiment. The bacterial communities were described using a molecular fingerprinting technique (terminal restriction fragment length polymorphism (tRFLP)), which allows descriptions of broad-scale changes in the taxonomic composition of these communities (Liu et al., 1997). tRFLP only identifies the most abundant taxa or those that are amplified most readily. All of the conclusions from the study therefore come with the caveat that they apply to what can be detected using tRFLP, whereas many more bacterial taxa undoubtedly occur in these communities.
Total nucleic acids were first extracted from microcosm communities (Bailey, 1995). Briefly, the samples were resuspended in 500 μl of lysis buffer containing 0.5% sodium dodecyl sulfate, 20 μg ml–1 proteinase K, 20 μg ml–1 of a mixture of lysing enzymes (Sigma, Gillingham, UK). The samples were left to stand at room temperature for 60 min, followed by incubation at 55 °C for 30 min. A solution containing 80 μl of 10% cetyl trimethylammonium bromide solution and 100 μl of 5 M NaCl was added and incubation continued at 65 °C for 10 min. Nucleic acids were extracted with 24:1 isoamyl alcohol:chloroform and precipitated with isopropanol. DNA pellets were cleaned with glacial ethanol, and suspended in 50 μl sterile deionized water. Bacteria-specific primers were used to amplify a roughly 500 bp locus. Each PCR reaction contained 25 pm D4 labelled primer 63f (5′-CAGGCCTAACACATGCAAGTC-3′) (Marchesi et al., 1998) and unlabelled 519r (5′- GTATTACCGCGGCTGCTG-3′) (Lane, 1991), 0.2 mM deoxynucleotide triphosphate (dNTPs), 1.25 U Taq DNA polymerase, 5 μg bovine serum albumin and 5 μl of 10 × PCR buffer in 50 μl volumes. PCR amplification consisted of an initial denaturing step at 94 °C for 1 min 30 s; 29 cycles of 94 °C for 45 s, 55 °C for 1 min and 72 °C for 1 min 30 s; final extension of 72 °C for 5 min. Millipore (Watford, UK) multiscreen filter plates were used to remove excess PCR reagents, and the remaining nucleic acids were resuspended in 25 μl deionized water. The nucleic acids were subjected to digestion in a 10 μl total reaction volume containing 10 U msp1 enzyme, 1 μl of 10 × enzyme buffer, 1 μg bovine serum albumin and approximately 50 ng PCR product. The reaction was incubated for 2 h at 37 °C and tRFLP profiles were analyzed on a Beckman CEQ2000 (Beckman coulter UK Ltd, Gillingham, UK) capillary sequencer. This produced a data matrix consisting of the relative abundances of detected peak sizes (species or operational taxonomic units) contained in each community. Peaks were assigned manually using GeneMarker 1.6 (SoftGenetics, State College, PA, USA) using the settings described by the company (Hulce and Liu, 2006).
Statistical analysis
This study attempts to explain dissimilarities between communities. I used the Bray–Curtis index to describe the pairwise dissimilarity between communities, in which the pairwise dissimilarity between sample i and j is  and where xik is the abundance of taxon k in sample i. The qualitative conclusions of this study were unchanged when different dissimilarity measures (Euclidean distance) or correlation (Pearson correlation) were used.
and where xik is the abundance of taxon k in sample i. The qualitative conclusions of this study were unchanged when different dissimilarity measures (Euclidean distance) or correlation (Pearson correlation) were used.
The study attempts to explain the degree of pairwise dissimilarity among communities in terms of several qualitative and quantitative explanatory variables. I use a multivariate version of the analysis of variance based on dissimilarities (Anderson, 2001) implemented in the adonis function of the vegan package (Oksanen et al., 2008) using R 2.8.1 (www.r-project.org). This non-parametric method partitions variation in dissimilarities into explained and unexplained portions in a manner analogous to analysis of variance. The reason the ‘adonis’ method was used rather than the more commonly used analysis of similarities (Clarke, 1993) is that adonis can accommodate both continuous and categorical predictors and their interactions. Mantel- and partial-Mantel tests, which examine correlations between dissimilarity matrices using randomization tests (Legendre, 1998), are frequently used for distance–decay studies, but produced the same qualitative results so are not reported here. In addition, Mantel tests could not be used for all of the analyses because some included categorical variables. All P-values are based on 999 permutations.
Results
Distance–decay in water-filled tree-holes
Surveys of water-filled tree-holes were conducted in Wytham Woods in 2008. Of the 14 surveys that were conducted in 2008, 2 were removed from the analysis because several of the DNA extracts could not be amplified. Of the remaining 12 surveys dates, 11 produced nonsignificant distance–decay relationships (adonis test; all P>0.1). There was a strong, positive relationship between community dissimilarity and geographic distance in the remaining survey (R2=0.41, adonis test P=0.005), which is marginally nonsignificant when the threshold P-value conservatively corrected for multiple tests using Bonferonni correction applied over the 12 tests (adjusted level=0.004). When the mean relative abundance of each taxon (tRFLP band) in each tree-hole was averaged over time, there was no evidence of a distance–decay relationship (adonis test P=0.16; Figure 1).
Distance–decay in microcosms
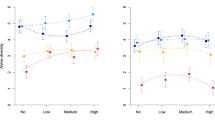
Microcosms containing sterile media were placed throughout the study area and colonization was allowed for 28 days. Of the 112 microcosms, 10 were either lost over the course of the experiment or did not yield DNA, leaving 102 microcosms (52 closed to colonization, 50 open to colonization). There was a significant positive relationship between geographic distance and community dissimilarity after 28 days in the microcosms that were open to colonization throughout (adonis test F1, 48=3.21, P=0.006, Figure 2a). The relationship seems to reach an asymptote in microcosms that are around 150 m apart, after which there is little increase in the degree of dissimilarity with increasing distance. The qualitative pattern of distance–decay was similar when the microcosms were closed to colonization (Figure 2b), and the relationship is also significant (adonis test F1, 50=6.71, P=0.001). As for the microcosms open to colonization throughout the 28 days (Figure 2a), the slope of the distance–decay relationship seems to decline when microcosms were around 150 m apart. An analysis of covariance using the mean dissimilarities at each distance indicated that the slope of the distance–decay relationship was marginally higher in the closed microcosms (F2, 12=4.6, P=0.03). When the raw dissimilarities were analyzed, there was a significant main effect of geographic distance (adonis test, F1, 98=7.00, P<0.001) but not of whether the microcosms were open to colonization (adonis test, F1, 98=1.41, P=0.16). There was, however, a marginally significant interaction between geographic location and whether or not the microcosm was closed to colonization (distance × colonization: F1, 98=1.81, P=0.048), confirming that the slope of the distance–decay pattern was steeper in microcosms that were closed to colonization over the last 14 days.
Distance–decay relationships in microcosms placed in Wytham Woods. Pairwise dissimilarities (Bray–Curtis index) are plotted as a function of the distance between the microcosms. Bacterial communities are described using tRFLP. Microcosms were left open to colonization for 2 weeks. The communities were then sampled after a further 2-week period during which the communities were (a) open to further colonization (b) closed to colonization. There was a significant relationship between geographic distance and community dissimilarity for both plots (adonis test, P<0.001).
Reciprocal transplants
Some of the microcosms at the extremities of the study area were transplanted to the other end of the study area. Of the 80 microcosms, 12 microcosms were lost or the DNA was not successfully amplified, leaving 68 microcosms. Within the set of the transplanted and the non-transplanted microcosms, the mean Bray–Curtis dissimilarity was approximately 0.58. Mean dissimilarity was higher (mean dissimilarity=0.74) when the transplanted microcosms were compared with the microcosms from their site of origin, indicating that the transplantation affected community composition.
The following factors were entered into an adonis model to assess their effect on dissimilarity among the community composition: (1) site (location within the study area); (2) whether or not the microcosm was transplanted to the other site; (3) whether or not the microcosm was closed to colonization; and (4) all interactions among these treatments (Table 1). The analysis found a significant effect of site (F1, 60=5.26, P<0.001), thus supporting the results of the microcosm distance–decay experiment (above) that spatial location explains variation in community composition.
In the transplantation experiment, dispersal limitation would be implied if the effect of transplanting the microcosm depended on whether or not the microcosm was closed to colonization (that is, if there is a significant colonization × transplantation interaction). The lack of a significant interaction term (Table 1) therefore provides evidence that dispersal limitation is unimportant in these experiments. Transplanting the microcosms altered the communities whether or not they were open to colonization (Table 1, P<0.001), indicating that simply putting the microcosm in a new location altered the communities. Since this did not depend on whether or not the microcosms were open to colonization, the only remaining possibility is that there must be some environmental factor that is affecting the communities. The results therefore provide little evidence for dispersal limitation, but that the communities are significantly altered when they are moved to a new location.
Manipulation of colonization rate
Colonization rate was manipulated by opening and closing microcosms. Of the 120 microcosms, 1 did not yield DNA and so was excluded from the analysis. There was an overall negative relationship between colonization rate and community dissimilarity (Figure 3). The steep decline in community similarity with increasing colonization rate was highly significant (F1, 117=4.24, P=0.0001, R2=0.04). The data for colonization rates of 2 and 6 days seemed to be driving the pattern. When these were removed from the analysis, the negative relationship remained highly significant (F1, 97=4.80, P=0.0001, R2=0.05) although the relationship was not as steep. Overall, it would seem that a few days is sufficient time period for most colonizing bacteria to arrive in the microcosms.
Discussion
The turnover of bacterial taxa over space and time is central to understanding their ecology and evolution. I showed that there was little evidence of a distance–decay relationship in the water-filled tree-holes studied here, at least at the spatial scales used in this study. In this respect, the study replicated the results of a number of recent surveys of microbial communities by showing that natural bacterial communities have the potential to produce distance–decay relationships, but that these tend to be weak (that is, shallow slopes) or non-existent over scales of metres to hundreds of metres (Horner-Devine et al., 2004).
Much of the variation in dissimilarity remains unexplained for the surveys of natural communities. One possibility is that community composition was determined principally by environmental factors that did not have a spatial pattern. For example, a previous study has shown that the volume of these communities has a pivotal role in determining taxonomic diversity (Bell et al., 2005), which did not vary systematically over the study area. Alternatively, the survey might have been conducted at an inappropriate spatial scale. For example, local adaptation of bacterial populations have been shown to occur at the scale of a few centimetres in soil (Vos et al., 2009) as well as at scales of tens to hundreds of metres (Belotte et al., 2003). Some observational surveys have only discovered significant distance–decay patterns once the surveys have been extended over hundreds of kilometres (Whitaker et al., 2003; Green et al., 2004) and the same might be true for the tree-hole ecosystem. Similarly, using more variable genetic loci to identify bacterial taxa might have identified variants that were dispersal limited (see below).
In spite of the lack of a distance–decay relationship in the natural tree-holes, there was considerable variation in community composition in the bacterial communities. Although there might be a spatial component that explains some of the variation in composition, there was no straightforward effect of geographic distance on community dissimilarity. Even if geographic distance was responsible for explaining some of the variation in community composition, these observational data are insufficient to identify the mechanisms that determine variation in composition. Several experimental manipulations were conducted to overcome the difficulties involved in conducting purely observational studies. The first manipulation placed microcosms containing sterile media in the woodland in which the observational surveys were conducted, thus attempting to minimize any environmental differences over space. The manipulation resulted in a highly significant distance–decay relationship, but could not identify the mechanisms that created the pattern. Assuming that environmental conditions were identical across the study area, the result indicates that dispersal limitation is acting on these communities, and that distance between microcosms was significant in determining the composition of the tree-hole communities in the absence of environmental differences. However, although the initial habitat was the same within each microcosm, there was the opportunity for environmental differences to arise across the study area, for example, if there was a subtle gradient of temperature or light, a possibility that could not be excluded from this experiment alone. Microcosms that were more exposed to sunlight might have both had warmer temperatures and allowed for the development of significant autotroph communities. Both of these factors would be expected to have considerable implications for bacterial community composition.
The distance–decay relationship became marginally more pronounced when the microcosms were closed to colonization for the final 2 weeks. This result shows that the distance–decay relationship persisted even when colonization was completely cut off. If the observed pattern was solely the result of continuing colonization (for example, community dynamics were neutral), it seems plausible that the distance–decay relationship would be expected to deteriorate as nearby communities became as different as distant communities in the absence of further colonization. One possible explanation is that there was an unobserved environmental gradient across the study area. If such a gradient existed, halting colonization might have allowed locally adapted individuals to increase in abundance and would therefore have produced a distance–decay relationship. Another possibility is that there were priority effects in combination with some degree of dispersal limitation, such that the first to arrive were able to exclude later colonists, and the pattern persisted even in the absence of further colonization. So far as I am aware, the only way to test these competing hypotheses is to conduct transplant experiments that simultaneously cut off colonization into the communities and alter environmental conditions by transplanting whole communities. I conducted a reciprocal transplant experiment to distinguish between these possibilities. As expected, there was greater similarity within communities at the same site compared with similarity between communities at different sites and microcosms transplanted to a new site took on the characteristics of communities at the new site. Unexpectedly, there was no effect on between-community dissimilarity of closing transplanted microcosms to colonization. The result implies that there were subtle environmental differences within the woodland that were responsible for driving community composition at least at these sites. On the basis of these results, of the possible hypotheses to explain the distance–decay relationship in initially sterile microcosms, the most likely would seem to be that there was an unobserved gradient in environmental conditions across the woodland. Certainly, the results of experiment provide little evidence of dispersal limitation at least over these spatial and temporal scales.
Another way of testing for dispersal limitation is to artificially mediate the amount of time that microcosms are open to colonization. I conducted such tests in microcosms at a single location by opening and closing microcosms to colonization at 2-day intervals over 30 days. There was a tendency for microcosms that were only open for a few days to be more dissimilar to the other communities compared with microcosms that were open to colonization for longer than a few days (Figure 3). The result suggests that any dispersal limitation would have been acting over a few days, but that most of the potential colonists would reach the microcosms over longer time scales. For the other experiments (Figure 2, Table 1), colonization was allowed into the microcosms for at least 2 weeks, so it would be anticipated that all colonists would have the opportunity to reach every microcosm over that time scale.
The combination of experimental and observational data allows a consistent picture of the microcosm communities to emerge. There was a clear distance–decay pattern in the microcosms which was maintained in microcosms closed to colonization (Figure 2). Transplanting the microcosms to new locations altered the communities, and this alteration was unaffected by closing the microcosms. The result provides a transparent demonstration of an environmental affect on the communities, and little evidence for distance dispersal, at least over these temporal and spatial scales. Finally, altering the colonization rate affected community dissimilarity over a few days, but was little affected after microcosms were open to colonization for longer than around 6 days. The temporal scale of the dispersal limitation was therefore likely to be over a few days, which might explain in part why dispersal limitation was not detected in the transplant experiment.
The molecular method used here (tRFLP) to describe the bacterial communities provides only a coarse description of these communities. Only the most abundant species are documented using this technique, and the patterns observed here might not hold for the many more taxa that were not detected using this method (Blackwood et al., 2007). The technique also suffers from some significant caveats, including sampling bias during the polymerase chain reaction, and the possibility that a single taxon produces multiple restriction fragment sizes (for example, Lueders and Friedrich, 2003). It is possible that the biases inherent in tRFLP resulted in conservative estimates of dispersal limitation. For example, it is plausible that the rarest taxa are those that are the most dispersal limited. The inability of the experiments to detect significant dispersal limitation might therefore reflect the inability of tRFLP to detect rare taxa rather than the importance of dispersal limitation in these communities. Future work will undoubtedly entail more fine-scaled efforts to estimate dispersal limitation for both common and rare taxa. Nonetheless, the consistent differences observed here in the tRFLP profiles reflect real differences in the composition or distribution of abundance in these communities. Another important caveat to the study is that the surveys and field experiments were conducted within a single woodland. Considerable research is still required to analyze whether the results apply to bacterial communities more generally or to tree-hole communities in other woodlands.
Assessing the importance of environmental differences on distance–decay relationships by measuring environmental variables (Green et al., 2004; Horner-Devine et al., 2004; Legendre et al., 2005; Ramette and Tiedje, 2007; Van der Gucht et al., 2007; Sommaruga and Casamayor, 2009) requires a good understanding of how the constituent species of complex microbial communities perceive the world around them. Although some environmental variables seem to have large effects on most bacteria species, such as pH (Fierer and Jackson, 2006), there remains an enormous amount of unexplained variation in bacterial community composition. One of the great advantages of studying bacterial communities is that whole communities can be manipulated under relatively natural conditions. In situ manipulations of bacterial communities, such as the transplant experiments described in this study, have the potential to be useful tools for understanding how environmental conditions influence the distribution and abundance of bacterial taxa. Overall, the experimental results here suggest that subtle environmental differences can quickly overcome dispersal limitation operating over time scales of a few days and at spatial scales of a few hundred metres.
References
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Bailey MJ . (1995). Extraction of DNA from the phylosphere. In: Trevors J, van Elsas JD (eds). Nucleic Acids in the Environment. Springer-Verlag: Berlin. pp 89–109.
Bell G . (2001). Neutral macroecology. Science 293: 2413–2418.
Bell T, Ager D, Song J-I, Newman JA, Thompson IP, Lilley AK et al. (2005). Larger islands house more bacterial taxa. Science 308: 1884.
Belotte D, Curien JB, Maclean RC, Bell G . (2003). An experimental test of local adaptation in soil bacteria. Evolution 57: 27–36.
Blackwood CB, Hudleston D, Zak DR, Buyer JS . (2007). Interpreting ecological diversity indices applied to terminal restriction fragment length polymorphism data: insights from simulated microbial communities. Appl Environ Microbiol 73: 5276.
Bradshaw WE, Holzapfel CM . (1992). Resource limitation, habitat segregation, and species interactions of British tree-hole mosquitos in nature. Oecologia 90: 227–237.
Carpenter SR . (1996). Microcosm experiments have limited relevance for community and ecosystem ecology. Ecology 77: 677–680.
Cho J-C, Tiedje JM . (2000). Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol 66: 5448–5456.
Clarke KR . (1993). Non-parametric multivariate analyses of changes in community structure. Austral Ecol 18: 117–143.
Fierer N, Bradford MA, Jackson RB . (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631.
Franklin RB, Mills AL . (2003). Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol Ecol 44: 335–346.
Green J, Bohannan BJM . (2006). Spatial scaling of microbial biodiversity. Trends Ecol Evol 21: 501–507.
Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M et al. (2004). Spatial scaling of microbial eukaryotic diversity. Nature 432: 747–750.
Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM . (2004). A taxa-area relationship for bacteria. Nature 432: 750–753.
Hubbell SP . (2001). The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32). Princeton University Press: Princeton, USA.
Hulce D, Liu C . (2006). GeneMarker software for terminal-restriction fragment length polymorphism (T-RFLP) data analysis. http://www.softgenetics.com/T-RFLPapplicationnote.pdf.
Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF . (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320: 1081.
Jessup CM, Forde SE, Bohannan BJM . (2005). Microbial experimental systems in ecology. Adv Ecol Res 37: 273–307.
Jessup CM, Kassen R, Forde SE, Kerr B, Buckling A, Rainey PB et al. (2004). Big questions, small worlds: microbial model systems in ecology. Trends Ecol Evol 19: 189–197.
Kitching RL . (1971). An ecological study of water-filled tree-holes and their position in the woodland ecosystem. J Anim Ecol 40: 281.
Lane DL . (1991). 16S/23S rRNA sequencing. In: Stackebrandt ER, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons: Chichester, UK, pp 115–175.
Legendre L . (1998). Numerical Ecology. Elsevier Science: Amsterdam, Netherlands.
Legendre P, Borcard D, Peres-Neto PR . (2005). Analyzing beta diversity: partitioning the spatial variation of community composition data. Ecol Monogr 75: 435–450.
Liu WT, Marsh TL, Cheng H, Forney LJ . (1997). Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63: 4516–4522.
Lueders T, Friedrich MW . (2003). Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl Environ Microbiol 69: 320–326.
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ et al. (1998). Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64: 795–799.
Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112.
Morlon H, Chuyong G, Condit R, Hubbell S, Kenfack D, Thomas D et al. (2008). A general framework for the distance–decay of similarity in ecological communities. Ecol Lett 11: 904.
Nekola JC, White PS . (1999). The distance decay of similarity in biogeography and ecology. J Biogeogr 26: 867–878.
Oksanen J, Kindt R, Legendre P, O’Hara B, Simpson BH, Solymos P et al. (2008). Vegan: community ecology package. R package version 1.15-1. http://cran.r-project.org/, http://vegan.r-forge.r-project.org/.
Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
Ramette A, Tiedje JM . (2007). Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci USA 104: 2761.
Sommaruga R, Casamayor EO . (2009). Bacterial’cosmopolitanism’and importance of local environmental factors for community composition in remote high-altitude lakes. Freshw Biol 54: 994–1005.
Srivastava DS, Kolasa J, Bengtsson J, Gonzalez A, Lawler SP, Miller TE et al. (2004). Are natural microcosms useful model systems for ecology? Trends Ecol Evol 19: 379–384.
Srivastava DS, Lawton JH . (1998). Why more productive sites have more species: an experimental test of theory using tree-hole communities. Am Nat 152: 510–529.
Suen G, Goldman BS, Welch RD . (2007). Predicting prokaryotic ecological niches using genome sequence analysis. PLoS ONE 2: e743.
Van der Gucht K, Cottenie K, Muylaert K, Vloemans N, Cousin S, Declerck S et al. (2007). The power of species sorting: Local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci USA 104: 20404.
Vos M, Birkett PJ, Birch E, Griffiths RI, Buckling A . (2009). Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325: 833.
Whitaker RJ, Grogan DW, Taylor JW . (2003). Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301: 976–978.
Woodcock S, van der Gast CJ, Bell T, Lunn M, Curtis TP, Head IM et al. (2007). Neutral assembly of bacterial communities. FEMS Microbiol Ecol 62: 171–180.
Acknowledgements
The research was supported by the Natural Environment Research Council grant NE/F000286/1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bell, T. Experimental tests of the bacterial distance–decay relationship. ISME J 4, 1357–1365 (2010). https://doi.org/10.1038/ismej.2010.77
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.77
Keywords
This article is cited by
-
Climate and Environmental Variables Drive Stream Biofilm Bacterial and Fungal Diversity on Tropical Mountainsides
Microbial Ecology (2024)
-
Spatial distribution pattern across multiple microbial groups along an environmental stress gradient in tobacco soil
Annals of Microbiology (2023)
-
Dispersal changes soil bacterial interactions with fungal wood decomposition
ISME Communications (2023)
-
Bacterioplankton Assembly Along a Eutrophication Gradient Is Mainly Structured by Environmental Filtering, Including Indirect Effects of Phytoplankton Composition
Microbial Ecology (2023)
-
Routes and rates of bacterial dispersal impact surface soil microbiome composition and functioning
The ISME Journal (2022)