Abstract
Most of the Shewanella species contain two periplasmic nitrate reductases (NAP-α and NAP-β), which is a unique feature of this genus. In the present study, the physiological function and evolutionary relationship of the two NAP systems were studied in the deep-sea bacterium Shewanella piezotolerans WP3. Both of the WP3 nap gene clusters: nap-α (napD1A1B1C) and nap-β (napD2A2B2) were shown to be involved in nitrate respiration. Phylogenetic analyses suggest that NAP-β originated earlier than NAP-α. Tetraheme cytochromes NapC and CymA were found to be the major electron deliver proteins, and CymA also served as a sole electron transporter towards nitrite reductase. Interestingly, a ΔnapA2 mutant with the single functional NAP-α system showed better growth than the wild-type strain, when grown in nitrate medium, and it had a selective advantage to the wild-type strain. On the basis of these results, we proposed the evolution direction of nitrate respiration system in Shewanella: from a single NAP-β to NAP-β and NAP-α both, followed by the evolution to a single NAP-α. Moreover, the data presented here will be very useful for the designed engineering of Shewanella for more efficient respiring capabilities for environmental bioremediation.
Similar content being viewed by others
Introduction
Nitrate is the most preferable substrate for bacterial anaerobic respiration because of its high redox potential. There are three types of prokaryotic nitrate reductase comprising of the cytoplasmic assimilatory (NAS), membrane-bound respiratory (NAR) and periplasmic dissimilatory (NAP) nitrate reductase used for nitrate respiration (Moreno-Vivian et al., 1999, Richardson et al., 2001, Stolz and Basu, 2002). All of them contain a molybdenum cofactor at their active sites; however their location, structure, sequence and terminal products are different. The distribution and physiological functions of NAS, NAR and NAP are different in different organisms. Many bacteria can express not only one functionally nitrate reductases, and they usually have important physiological roles. For example, Escherichia coli harbors two NARs and one NAP. The first NAR serves at high nitrate concentration (Potter et al., 1999), the second NAR is expressed in response to stress and is part of the RpoS regulon (Spector et al., 1999); and the NAP enzyme is primarily expressed at low nitrate concentrations, and assists the nitrate respiration via NAR, in which bioenergetic efficiency is limited (Stewart et al., 2002). Paracoccus pantaotrophus contains NAS, NAR and NAP, and each of them serves different functions (Sears et al., 1997, Tabata et al., 2005).
At present, the availability of a large number of microbial genomes has made it possible for us to get insight view of the distribution and evolution of prokaryotic nitrate reductase. A recent review of the genomes of 19 Shewanella strains shows that most of the Shewanella contain two types of NAP: NAP-α and NAP-β (Simpson et al., 2010). Considering the unparalleled capability of Shewanella to utilize a variety of substrates, the presence of two NAPs may reflect an adaptation of Shewanella to cope with the various environmental conditions. There are several implications for this special feature of Shewanella genus: (1) The excess nap gene cluster may not be expressed, similarly as the two dimethylsulfoxide reductase gene clusters in S. oneidensis MR-1 (Gralnick et al., 2006); (2) The two NAP systems may have distinct physiological functions such as NAP-α may serve in denitrification or redox balancing (Simpson et al., 2010); (3) The double copy of nap gene are functionally redundant. Besides all these possibilities, it is still an open question that what is the evolutionary relationship of the two NAPs? And why it happens only in Shewanella genus?
Generally the basic components of NAP are napDABC. The cytoplasmic protein NapD is involved in NapA relocation by binding to the NapA twin-arginine signal peptide (Maillard et al., 2007). The NapAB form heteromeric complex catalyses by the reduction of nitrate to nitrite. The electrons from quinol pool are generally passed through one or two cytochrome-c containing proteins (NapC and NapB (di-c-haem cytochrome redox partner of NapA)) to the catalytic subunit, NapA that contains a Mo-bis-molybdopterin guanine dinucleotide cofactor and a cluster. The NapC is a tetrahaem c-type cytochrome of the NapC/NirT family, consists of ∼175 amino acid residues that are assumed to form an N-terminal membrane-spanning helix and a globular cytochrome-c domain situated at the outside of the bacterial membrane (Gross et al., 2005).
A review of 136 species across 51 genera shows that most of the nap gene clusters contain napC. In the napC free strains, such as Campylobacter jejuni and Wolinella succinogenes, NapGH are at the substitution place of NapC (Pittman et al., 2007, Kern and Simon, 2008). In E. coli, which contains NapGH and NapC, NapGH is not involved in menaquinol oxidation and instead form a quinol dehydrogenase that transfers electrons from ubiquinol through NapC to NapAB (Brondijk et al., 2002, 2004). In the well-studied S. oneidensis MR-1, which also doesn’t have a napC gene, CymA has key roles in both nitrate and nitrite reduction (Murphy and Saltikov, 2007; Gao et al., 2009). CymA is also a tetrahaem c-type cytochrome belonging to NapC/NirT family, and it has been proven to be the essential electron transport protein in 4–6 pathways studied in Shewanella (Myers and Myers, 2000, Murphy and Saltikov, 2007). As most of the Shewanella strains contain both a napC dependent and a napC independent nap gene clusters, it is crucial to study and distinguish the roles of NapC and CymA in the electron transport in Shewanella.
In the present study, we investigated the two NAP systems in Shewanella by using a combination of bioinformatics, genetic and phylogenetic analyses using S. piezotolerans WP3 as a model organism. The strain WP3 was isolated from 1.9 km depth marine sediment in west Pacific Ocean and its whole genome has been sequenced (Xiao et al., 2007, Wang et al., 2008). Two nap gene operons are identified in WP3 genome: napDABC (swp_2775-2772) and napDAB (nap_4456-4458). Here, we show that both NAPs are functional in anaerobic nitrate respiration, and NAP-α confers a selective advantage to WP3 in nitrate utilization. On the basis of the data presented, we have proposed an evolutionary scenario of NAPs in Shewanella: NAP-β as the prototype at beginning, then NAP-α was obtained to form the dual NAP system containing both NAP-α and NAP-β, and finally would evolve to a single NAP-α.
Materials and methods
Bacterial strains, media and culture conditions
All bacterial strains and plasmids used in the present study are listed in Table 1. The Shewanella strains were cultured in marine broth medium 2216E at 20 °C. The E. coli strain WM3064 was incubated in Luria-Bertani medium (Trypton: 10 gl−1, yeast extract: 5 gl−1, NaCl: 10 gl−1) at 37 °C. Chloramphenicol (25 μg ml−1 for E. coli; 12.5 μg ml−1 for WP3) was added to the medium when required.
A modified 2216E medium (Trypton: 5.0 gl−1, Yeast extract: 1.0 gl−1, NaCl 34 gl−1, D-L lactate: 20 mM, pH=7.0) supplemented with sodium nitrate was used for physiological characterization of the mutant under anaerobic condition. The OD600 was measured using a SHIMADZU UV-2550 spectrophotometer (SHIMADZU CO., Kyoto, Japan) to draw growth curve. The LML medium (Cruz-Garcia et al., 2007) was also used in case.
For anaerobic cultivation, a modified method was used (Miller and Wolin, 1974; Balch and Wolfe, 1976). Briefly, media that didn’t contain any electron acceptor were prepared under non-sterile condition. Then the media were dispensed into serum bottles gassed with O2-free nitrogen, and the stoppers were inserted as the bottles were withdrawn from gassing needles. Metal seals were then crimped to seal the caps, and then the media were autoclaved with a needle inserted into the stopper. When the autoclave was done, the needles were plucked off immediately. After the media were cooled, the serum bottles were gassed by a gassing manifold system (Balch and Wolfe, 1976). Nitrate solution (0.4 M, sodium nitrate) was filter sterilized in a vinyl anaerobic airlock chamber (Coy Laboratory Products Inc., Grass Lake, MI, USA) and added to the concentration needed. The serum bottles, stoppers and metal seals were bought from Wheaton Science Products (Millville, NJ, USA).
Development of the mutant strains
The genes napA1, napA2, napC and cymA were deleted in-frame using a fusion PCR method with plasmid pRE112 as described earlier (Wang et al., 2009). The primers used for generating PCR products for mutagenesis are listed in Table 2. To construct a napA1 in-frame deletion mutant, two fragments flanking napA1 were amplified by PCR with primers swp2774up1 and swp2774up2, primers swp2774do1 and swp2774do2, respectively. The fusion PCR products were generated by using the amplified fragments as templates with primers swp2774up1 and swp2774do2, and ligated into the Sac I and Xba I site of plasmid pRE112. This resulted in plasmid pRE112-napA1 and was first transformed into E. coli WM3064 by calcium chloride transformation and then into WP3 by conjugation. Integration of the mutated version of napA1 into the chromosome was selected by chloramphenicol resistance and further confirmed by PCR. The verified transconjugants were grown in 2216E broth in the absence of NaCl and plated on 2216E medium supplemented with 10% of sucrose. The chloramphenicol-sensitive and sucrose-resistant colonies were screened by PCR for the deletion of napA1.
The same strategy was used for constructing napA2, napC and cymA in-frame deletion mutants with primers listed in Table 2. Double mutant ΔnapA1ΔnapA2 was constructed either by introducing pRE112-napA2 into the ΔnapA1 or by introducing pRE112-napA1 into the ΔnapA2. Same strategy was used to construct ΔnapA1ΔcymA, ΔnapA2ΔnapC and ΔnapCΔcyma, respectively.
Complementation assay of the mutant
For complementation, we used a Shewanella–E. coli shuttle plasmid vector pSW2 developed from the filamentous phage SW1 of WP3 (Wang et al., 2007, our unpublished data). To introduce cymA gene into the ΔcymA mutant, cymA along with its native promoter region was amplified by PCR using WP3 genomic DNA as the template and primers RC-cymA-F and RC-cymA-R as listed in Table 2. The PCR product and pSW2 were digested with both BamH I and Xho I, respectively, and then ligated to generate pSW2-cymA. The plasmid was introduced into WM3064 by calcium chloride transformation then mobilized into ΔcymA by mating. The same strategy was used to construct complementing strain for ΔnapCΔcymA.
RNA extraction and reverse transcription
The total RNA from cells was extracted as described previously (Wang et al., 2009). Total RNA was isolated from the mid-log phase (OD600≈0.2) cells culture growing at different conditions with TRI reagent-RNA/DNA/protein isolation kit (Molecular Research Center Inc., Cincinnati, OH, USA) following the manufacturer's instructions. The RNA samples were treated with DNase-I at 37 °C for 1 h and then purified with RNeasy Mini Kit (Qiagen, Hilden, Germany). The purified RNA samples were used to synthesize complementary DNA with the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Burlington, ON, Canada) following the manufacturer's instructions.
Real-time PCR
The primer pairs (Table 2) for the selected genes for real-time PCR were designed using Primer Express software (Applied Biosystems, Foster City, CA, USA). The PCR was performed using 7500 System SDS software in reaction mixtures with total volumes of 20 μl containing 10 μl SYBR green-I Universal PCR Master Mix (Applied Biosystems, Warrington, UK), 0.5 μM each primer and 1 μl complementary DNA template. The amount of target was normalized to that of the reference gene swp_2079, whose expression remains stable under various conditions relative to the calibrator (the transcription levels of the genes at aerobic condition, nitrate free medium, 20 °C, were set as 1). The real-time reverse transcription-PCR assays were performed in triplicate for each sample, and a mean value and standard deviation were calculated for the relative RNA expression levels.
Determination of nitrite concentrations in culture
The 5 μl of the culture supernatants from anaerobic growth experiments were diluted to 250 μl with deionized water, then added with 500 μl 1% (w/v) sulphanilamide (Bio Basic Inc., Markham, ON, Canada) dissolved in 25% (v/v) HCl and 500 μl of 0.2% (w/v) 1-naphthylamine dissolved in 25% (v/v) acetic acid, successively. After 40 min, the absorbance at 540 nm was measured using a SHIMADZU UV-2550 spectrophotometer, and nitrite concentrations were determined in comparison with the standard curve.
Competition assay
The competition assay was performed as described previously (Gao et al., 2009). Anaerobic cultures of wild type and ΔnapA2 were grown independently to stationary phase in modified 2216E medium supplemented with 2 mM sodium nitrate to OD600 at 0.3. And competition assay was performed in 4 mM nitrate concentration. Primers used for scanning were listed in the Table 2. Relative fitness, W, was calculated as described earlier (Lenski et al., 1991).
Constructing the phylogenetic tree
The phylogenetic tree (Figure 1) contains 37 NapA protein sequences from 21 genomes of Shewanella species. The phylogenetic tree in Figure 2 contains 183 NapA sequences, which are mainly from those previously described (Simpson et al., 2010), in addition we added the NapA of S. violacea DSS12, S. benthica KT99, and monomeric NAS of Bacillus subtilis, Klebsiella oxytoca, Rhodobacter capsulatus and Synechococcus elongatus. We used formate dehydrogenase protein of Bacillus cereus as the out-group, as formate dehydrogenase also belongs to DMSO (dimethyl sulfoxide) reductase family protein. The protein sequences were multiply aligned by ClustalW (Thompson et al., 1999), using BLOSUM matrix, with the gap open cost 7 and gap extend cost 0.1. Then posterior tree was performed by the methods of MrBayes (Huelsenbeck and Ronquist, 2001) with equal rate variation. The posterior probabilities were used as measures of statistical support for the branches. All these processes were performed by using the software Geneious 4.8.3 (Biomatters Ltd., Auckland, New Zealand) (Drummond et al., 2010) along with the ClustalW 2 and the Mrbayes 3.1.2. GenPept accession numbers of NapA protein sequences used for Figures 1 and 2 and are listed in Supplementary information Table S1.
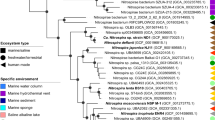
Phylogenetic tree of bacterial NAPs. The tree contains 185 NapA protein sequences, which come from the genomes of 165 prokaryotes across 53 genera and 4 monomeric Nas. We use formate reductase of Bacillus as out-group using Mrbayes method. Name abbreviations: Cam, Campylobacter jejuni; Wol, Wolinella succinogenes; Sul, Sulfurovum; Nau, Nautilia profundicola; Kle, monomeric Nas of Klebsiella oxytoca; Des, monomeric Nap of Desulfovibrio desulfuricans; Din, Dinoroseobacter shibae; Dec, Dechloromonas aromatica; Mag, Magnetospirillum magneticum; Hae, Haemophilus; Man, Mannheimia succiniciproducens; Ser, Serratia proteamaculans; Yer, Yersinia; Pec, Pectobacterium atrosepticum; Cit, Citrobacter koseri; Sal, Salmonella; Esc, Escherichia; Shi, Shigella; Aer, Aeromonas; Pho, Photobacterium profundum; She Nap-α-1, Nap-α-1 group of Shewanella genus; Vib, Vibrio; She Nap-α-2, Nap-α-2 group of Shewanella genus; Sac, Saccharophagus degradans; Bra, Bradyrhizobium sp.; Pse, Pseudomonas; Azo, Azoarcus ap. BH72; Bor, Bordetella bronchiseptica; Ral, Ralstonia; Sin, Sinorhizobium meliloti; Rho, Rhodospirillum centenum; She Nap-β, Nap-β group of Shewanella genus.
Results
Evolutionary relationships of NAPs
A phylogenetic tree of NapA proteins from 21 genomic sequences of Shewanella (sequence obtained from NCBI GenPept) was constructed as shown in Figure 1. On the basis of the phylogenetic analysis, the NapA orthologous in the genus Shewanella could be divided into NAP-β and NAP-α as suggested previously (Simpson et al., 2010). The NAP-α cluster could be further divided into two subclusters: Nap-α-1 and Nap-α-2. The Nap-α-1 group includes NAPs from S. violacea, S. benthica, S. sediminis and S. piezotolerans, all of which are psychrophilic/psychrotrophic. In the genomes of the compared Shewanella strains, dual NAP system consisting of NAP-β and either Nap-α-1 or Nap-α-2 is the major genotype, although sole NAP-β (S. oneidensis and S. halifaxensis HAW-EB4) or sole NAP-α (S. denitrificans, S. violacea and S. benthica) is the minor genotype.
The evolutionary relationships of Shewanella NAPs and other known NAPs were carefully analyzed by constructing a robust phylogenetic tree containing 186 NapA proteins from 165 different organisms of 53 genera (Figure 2). The tree has three major branches: ‘Branch 1’ is comprised of NAP-β including the monomeric NAP subclass that does not contain NapB, but instead contains a NapM, NapG or predicted protein as redox partners. ‘Branch 2’ is generally comprised of NAP-β from enterobacteriaceae family bacteria, and only the members near Yersinia belong to NAP-α. ‘Branch 3’ has large population of NAP-α, with only NAP-β of Shewanella and Aeromonas nearby branch root. The tight NapAB interaction subgroup and monomeric NAP subgroup are highlighted in the tree. The tight NapAB interaction subgroup is widely accepted as a more evolved group, and monomeric NAP is the evolutionary link from NAS to NAP (Jepson et al., 2006). The Nap-α-1 of Shewanella forms a close cluster with those from Vibrio species, while the NAP-α-2 forms a relatively independent branch. It is also noticed that the NapC containing NAP accounts for a large proportion of NAP, while the NapC free NAP (Tagged with *) only contains two parts: the monomeric NAP with its nearby members and the NAP-β of Shewanella.
NAP operons in S. piezotolerans WP3
According to the original annotation of WP3 whole genome, there are two sets of genes encoding NAP: napDABC (swp_2772-2775) and napDAB (swp_4456-4458). We named them as WP3-nap-α(napD1A1B1C1) and WP3-nap-β(napD2A2B2) (Figure 3). Besides NAP, none of the other nitrate reductase was identified in WP3. Five putative nitrite reductase (nrfA) genes were identified: swp_3958, swp_0919, swp3403, swp_0614 and swp_0613, and no other genes belonging to nrf family were found. Moreover, the genome of WP3 was analyzed for the presence of NirS (cytochrome cd1 nitrite reductase) and NirK (copper-containing nitrite reductase), which catalyze nitrite reduction in denitrification pathway, but none of them were found in the genome. In addition, the gene encoding CymA tagged as swp_4806 was found. The blast (McGinnis and Madden, 2004) analyses showed that NapA1 protein sequences share 71% identities with NapA2 and, 94, 85 and 73% identities with that of NAP-α of S. sediminis HAW-EB3, Vibrionales bacterium and S. woodyi ATCC 51908, respectively.
Nitrate respiration in WP3
The 2216E medium supplemented with lactate and sodium nitrate was used to study the anaerobic nitrate respiration of WP3, the time course of nitrite concentration was monitored along with the cell density in the medium. The growth curve of WP3 in different nitrate concentration ranging from 0.5–6 mM is presented in Figure 4a. The cultures with 2 mM nitrate were shown to have the shortest time to reach the stationary phase, and the cultures with 4 mM nitrate showed highest cell density at stationary phase. Together with the time course curve (Figure 4b), it was observed that nitrite reached a maximum and then decreased due to its reduction to ammonia. The growth curve did not reach to the stationary phase until the nitrite was completely consumed. This phenomenon indicates that the toxin of nitrite will limit cell growth, and the reduction of nitrite is the major process to generate ATP during nitrate respiration.
Transcription assays of the two nap gene operons
To get a hint if the two NAP systems in WP3 are functional, transcriptional assays of the two nap operons were performed by reverse transcription-PCR. The cells were incubated in medium supplemented with 2 mM sodium nitrate and 20 mM lactate anaerobically at 20 °C for RNA extraction (see the Materials and methods). Meanwhile, the transcription patterns of the two NAP operons were also assessed at 4 °C in accord with the most common environmental temperature of the deep-sea sediment where WP3 was isolated. Both the WP3-nap-α and WP3-nap-β gene clusters were induced by nitrate, and the expression level of napA2B2 is much higher than that of napA1B1 (60 folds vs 2 folds induction) (Figure 5). Interestingly at 4 °C, napA1B1 was highly upregulated (10 folds) comparing with its expression at 20 °C, whereas the expression of napA2B2 was downregulated by low temperature. These data suggested that the two nap operons have different regulatory mechanisms.
Transcription levels of nap genes. The gene transcription of WP3 cells incubated in 2216E medium (2 mM sodium nitrate, anaerobic) was monitored by reverse transcription-PCR as described in the Materials and methods. The pattern of the columns represents different incubating conditions. All the data were normalized to 20 °C aerobic growth without nitrate. Data are averages of triplicate experiments.
Both NapA1 and NapA2 are functional
The napA genes were disrupted to confirm whether both of the nap gene operons are functional or not. The mutant ΔnapA1 or ΔnapA2 still showed nitrate-reducing ability, but the double deletion mutant ΔnapA1ΔnapA2 lost the ability of nitrate reduction and could not survive on nitrate (Figure 6a). Therefore, it was confirmed that both of the nap gene clusters are functional. In the medium with 2 mM sodium nitrate, the growth and time course nitrite concentration of ΔnapA1, ΔnapA2 and the wild-type WP3 seem to have no difference (Figures 6a and c). As the WP3 culture grown at 4 mM nitrate has shown the highest cell density at stationary phase (Figure 4a), the mutated strains were also incubated at nitrate concentration of 4 mM. Surprisingly, the ΔnapA2 strain showed faster growth than both the mutant ΔnapA1 and the WT, and it had higher cell density at stationary phase than the other two strains (Figure 6b). The nitrite in the culture of ΔnapA2 is also the first to exhaust (Figure 6d). However, nearly no difference was observed between the WT and the ΔnapA1 mutant, no matter on growth rate or in time course nitrite concentration. Similar results were obtained when the strains were grown in LML medium (Cruz-Garcia et al., 2007) with 4 mM sodium nitrate (data not shown). These results suggest that the genotype of ΔnapA2 is more adapted for nitrate respiration.
Growth of WP3 wild type and mutant strains on nitrate under anaerobic condition (a, b), and corresponding time course nitrite concentration (c, d). In all panels (•)ΔnapA1ΔnapA2; (▴)ΔnapC; (▪)ΔnapA2; (○)ΔnapA1 and (▵)WT are common. (a) Growth on 2 mM nitrate. In this panel, ΔnapA1ΔnapA2 cannot grow, and in (c) no nitrite was detected. In both (a, c) ΔnapA1=ΔnapA2=WT, (b) Growth on 4 mM nitrate and corresponding nitrite concentration (d). In both (b, d) ΔnapA1=WT. Data are averages of triplicate cultures.
ΔnapA2 has competitive advantage
A growth competition assay was performed between the mutant ΔnapA2 and the WT. It was observed that 90% of the growth was occupied by the mutant ΔnapA2 after five times transfer (Table 3). In T0 samples, the average number of colonies was 356, of which 49% were the mutant ΔnapA2 (100 colonies per plate were examined by colony PCR). After 1-day incubation, the number of the ΔnapA2 increased to 63%. After 5 days, the ΔnapA2 contributed up to 94 % of the total population. The relative fitness values of ΔnapA2 vs WT from T1 vs T0, T5 vs T0 and T5 vs T1 were 1.1301, 1.1335 and 1.1344, respectively. These data suggest that ΔnapA2 has a competitive advantage in nitrate respiration.
Role of NapC and CymA in nitrate utilization
The mutant strain ΔnapC showed growth deficiency as compared with the WT, ΔnapA2 and ΔnapA1, independent of nitrate concentrations (2 or 4 mM) (Figures 6a and b), and it took more time for the nitrite consumption (Figure 6d). The mutant ΔnapA1 and WT have shown no phenotypic difference when grown in nitrate medium. These data demonstrate that NapC is an indispensable part of NAP systems in WP3, the function of NapA1 could be fully compensated by NapA2, but the function of NapC cannot be fully compensated by other electron transport proteins.
As the WP3-nap-β gene cluster doesn’t have napC, so the CymA is supposed to have the role of NapC. A ΔcymA mutant and a ΔnapCΔcymA were developed and their phenotypes were observed (Figures 7a and b). The growth of ΔcymA mutant was significantly decreased in the nitrate media, and maximum growth could be measured up to OD600 0.1 whereas the cell density of wild-type WP3 could reach to 0.3 OD600 units. Meanwhile, the double gene mutant ΔnapCΔcymA showed very poor growth (cell density of 0.04 OD600 units), and traces of nitrite were detected in the culture. These data suggest that both NapC and CymA are involved in nitrate reduction, and CymA is the sole electron transfer protein between quinol pool and nitrite reductase. The cymA gene was cloned into plasmid pSW2 (a Shewanella–E. coli shuttle vector constructed in our lab, see Materials and methods) to perform complementation assay for the further confirmation of this phenomenon. It was observed that the ability of nitrite reduction of mutant ΔcymA was partially restored by the plasmid harboring cymA gene. When the same plasmid carrying the cymA gene was transformed into the double mutant ΔnapCΔcymA, the reducing abilities of both nitrate and nitrite were partially restored (data not shown).
(a) Growth of WP3 wild type and mutants on 2 mM sodium nitrate under anaerobic condition and (b) corresponding time course nitrite concentration curve. In both panels, (•)ΔnapCΔcymA, (▴)ΔnapA2ΔnapC, (▪)ΔnapA1ΔcymA, (○)WT, (▿)ΔcymA and (•)ΔnapA1ΔnapA2 are common. ΔcymA can reduce nitrate to nitrite, but cannot further reduce nitrite. The cell density of ΔcymAΔnapA1 sustains at a low level, and the nitrite accumulation is very slow. ΔnapCΔcymA only have a low level of cell density, and trace amount of nitrite can be detected in its culture. In both (a, b) WT=ΔnapA2ΔnapC. Data are averages of triplicate cultures.
Electron transport preference of NapC and CymA
To test whether NapA2 can acquire electron from NapC and whether CymA can transfer electron to NapA1, mutants ΔnapA1ΔcymA and ΔnapA2ΔnapC were developed. Interestingly, both the mutants were shown to retain the nitrate reducing ability (Figures 7a and b). It indicates that NapA1 and NapA2 can acquire electron for nitrate reduction from both NapC and CymA. However, the nitrate reducing ability of ΔnapA1ΔcymA mutant is much lower than the ΔcymA mutant (Figures 7a and b), which suggests that electron flow from NapC to NapA2B2 is not as efficient as from NapC to NapA1B1. On the other hand, the electron flow efficiency from CymA to NapA1B1 is nearly the same as that from CymA to NapA2B2. These data indicate that CymA transport electron to both NapABs unspecifically, and NapC prefers to transport electrons to NapA1B1.
Discussion
Electron transport chain in nitrate ammonification of WP3
Based on the above experimental data and current knowledge on nitrate respiration, an electron transport pathway of nitrate ammonification for WP3 is depicted at Figure 8a. The nitrate was reduced to ammonia by a two-step process, in which nitrite reduction would not start until nitrate was used up (Cruz-Garcia et al., 2007). As shown in the model, NapC and CymA transferred electrons from menaquinol to both NapA1B1 and NapA2B2, and NapC preferentially transfers electrons to NapA1B1. When nitrate was completely consumed, CymA delivers electrons to NrfA directly, and CymA is the sole electron transport protein to NrfA. Five putative nrfA genes were found in WP3 genome, but only one gene (swp_3958) was proven to have function in anaerobic nitrite reduction in WP3 (data not shown). In the mutant strain ΔnapA2, NapC (or maybe part of CymA also) is supposed to be able to fulfill the electron transfer function from quinol pool to NapA1B1, then CymA can transport freely available electrons to NrfA instantly after nitrate reduction to initiate nitrite reduction (Figure 8b). In the ΔnapC mutant, CymA should mediate the electron flow to NapAB, and then transfer electron to NrfA. As CymA is at the promiscuous place, the nitrite reduction is not as efficient as in the WT (Figure 8c). In the case of ΔnapA1, the electron flow from CymA to NapAB and then to NrfA was not been influenced, and the function of NapA1 could be fully compensated by NapA2, thus the loss of napA1 gene would not affect nitrate respiration (Figure 8d). We also observed that there was noticeable amount of nitrite generated in the culture of ΔnapCΔcymA mutant, which indicates that there is a third electron transport protein that has minor function in delivering the electrons to nitrate reductase. Different from S. oneidensis MR-1 that contains NapGH, no NapGH exist in WP3. We suppose that the protein TorC who also belongs to NapC/NirT protein family with similar function and structure, may take a minor function of NapC and CymA in WP3.
Electron transport model for nitrate ammonification. (a) In wild-type WP3, electrons transfer from menaquinol (MKH2) to periplasmic nitrate reductase NapAB by NapC and CymA. NapB serves as a direct electron transfer to NapA without catalytic function. When nitrate is exhausted electron will be transferred to nitrite reductase NrfA by CymA directly. (b) In the absence of NapA2, CymA can transfer electron to NrfA specially. (c) In ΔnapC mutant strain, nitrate reduction is less efficient than WT. (d) In ΔnapA1 mutant, because enzyme activity of NapA1 can be compensated by NapA2, the deletion of napA1 gene will not affect the growth. Solid lines between proteins represent electron flows. Dash lines in (a, d) indicate that electrons may be transferred from NapC to NapA2, this transferring pathway has been observed in the absence of NapA1 and CymA. Thin line in (c) indicates that the efficiency of this electron transfer may be reduced in the absence of NapC. Thick arrow in (b) indicates that the efficiency of the electron flow from this pathway may be higher in the mutant.
Why do most Shewanella contain two NAP systems?
As stated above, Shewanella is unique in having two sets of NAP systems, NAP-α with NapC and NAP-β without NapC. In E. coli, NapC was proved to be able to transfer electrons from both ubiquinol and menaquinol pool and it was considered that the activity of the NapC-based complex (NAP-α) or the NapGH-based complex (NAP-β) in Shewanella may be directed by the composition of the available quinol pool, which may depend on the presence of oxygen (Simpson et al., 2009). However, in our study, we found that both Nap-α and Nap-β contribute to anaerobic nitrate respiration of WP3. No nitrite was detected when WP3 incubated under aerobic condition with the medium added with nitrate. Both systems were found induced by nitrate under anaerobic condition, but not expressed at aerobic condition with nitrate. Now the question arises, why does Shewanella contain NAP systems with similar functions in anaerobic respiration?
In nature, the substances that can be respired by bacteria are changing in the environment, and usually several electron acceptors are accessible to microbes at the same time. Therefore, it is quite possible that possessing a set of gene, which is dedicated to a certain type of substance, will confer a selective advantage to the organism. The Shewanella species are famous for their respiratory versatility, and the electron transport protein CymA participates in many pathways such as metal, dimethylsulfoxide, nitrate and nitrite reduction in the studied Shewanella (Murphy and Saltikov 2007). The NapC, which transfer electron to NapAB specifically, is a dedicated electron transport protein. In the case of WP3, CymA was proven to be shared by Fe3+ reduction, nitrate and nitrite reduction (unpublished data and the present study). Therefore, containing both napC and cymA are better than containing cymA only. That is to say in nitrate ammonification electron transport to NapAB and NrfA by different transport protein could be more efficient than by a multi-functional protein only. Our data support the idea that Shewanella may use CymA as universal component for many electron transport pathways at beginning and obtained dedicated electron transfer proteins for some of these pathways with time (Gao et al., 2009). This would explain why most of the Shewanella contain two nap operons.
NAP-α is cold inducible
Although both NAP systems in WP3 contribute to anaerobic nitrate respiration, and both are induced by nitrate anaerobically. It is very interesting to notice that the NAP-α genes are highly induced at low temperature (4 °C) comparing with those at its optimal growth temperature (20 °C), while the transcription of NAP-β is relatively downregulated by low temperature (Figure 5). We also assessed the influence of high pressure to the nitrate respiration of WP3 (Wang et al., 2008 and our unpublished microarray data), however the expression of the two NAPs was not changed under different pressures. The induction of NAP-α at low temperature suggests that NAP-α may benefit the nitrate respiration of WP3 in the cold environments. Shewanella species are renowned for their capability to thrive at cold conditions. At low temperature, most of the biochemistry reactions become slow. To cope with this, cold living organisms may increase the concentrations of their enzymes, and/or improve their catalytic efficiencies at low temperature. Considering that NAP-α is a dedicated and more efficient nitrate reducing system, induction of NAP-α would be a wise strategy to respire nitrate at low temperature. We hypothesize that when WP3 lives at low temperature, it produces more NAP-α for more efficient energy production through nitrate respiration, as more free CymAs could be available to deliver electrons to NrfA for nitrite reduction. Therefore, it is an economic way to respire nitrate for energy production at cold conditions.
A binding motif for NarP was found in the upstream regions of all the NAP-α and NAP-β of Shewanella, suggesting their expression control under the NarP two-component regulation system. However, no typical cold shock box sequence was found in the potential promoter regions of the NAP genes (unpublished data). Interestingly, a gene encoding a DEAD box RNA helicases: HrpA is always located in the upstream of the NAP-α operons of the Shewanella (except S. denitrificans and S. frigidimarina). It is possible that this HrpA protein may regulate the transcription and facilitate the mRNA processing of NAP-α at low temperature. Containing a cold inducible NAP-α may be a widely utilized strategy for Shewanella genus to thrive at low temperature.
The evolution of two nap operons
The phylogenetic analyses of NAP systems showed that horizontal gene transfer of the NAP systems has frequently occurred among bacteria and NAP-β is a more ancient system in Shewanella than the Nap-α-1 and Nap-α-2 groups (Figures 1 and 2). The growth curve and the competition assay demonstrate that NAP-α confer a competition advantage to WP3 and possessing only Nap-α is better than possessing both NAP-α and NAP-β, or NAP-β only. Together with the fact that almost all Shewanella can use metal and many electron transport pathways share CymA as a common component. It is very likely that the CymA-dependent respiration system is more ancient. If it holds S. oneidensis MR-1, which only has NAP-β, may possess a preliminary anaerobic respiration system, whereas S. benthica and S. violacea, which contain only NAP-α may represent a more evolved respiration system. S. denitrificans also contains a single NAP-α, but it doesn’t contain cymA, its evolution process of respiration system may be different from most of the Shewanella. Here, we suppose whether the time is long enough for the NAP-β of some Shewanella, which will be eliminated through the selection, and there are some facts to support our hypothesis. In the genome of S. frigidimarina whose NAP-β is only comprised of napAB, and in S. baltica, S. woodyi and WP3 the NAP-β is napDAB. Considering the cold inducible property of NAP-α in WP3, and the psychrophilic property of S. benthica and S. violacea, cold may be one of the selection powers to make this process happen.
In conclusion, we have shown that the NAP-α and NAP-β systems in S. piezotolerns WP3 are both functional in anaerobic nitrate respiration. Both NapC and CymA are essential in electron transport for nitrate reduction. Containing two NAP systems in Shewanella genomes may be a reflection of the intermediate stage of evolution: it had a preliminary nitrate reduction system (NAP-β) sharing CymA with some other respiring systems at the beginning; then acquired a dedicated nitrate reducing system (NAP-α) with time for more efficient nitrate utilization; would evolve to a single NAP-α at the next step of evolution. However, we cannot exclude the possibility that NAP-β would be useful under certain conditions. Our study will give some hints for considering the evolution of other respiration systems in Shewanella and even in other bacteria, and will be very useful for the designed engineering of Shewanella for more efficient environmental bioremediation.
References
Balch WE, Wolfe RS . (1976). New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol 32: 781–791.
Brondijk THC, Fiegen D, Richardson DJ, Cole JA . (2002). Roles of NapF, NapG and NapH, subunits of the Escherichia coli periplasmic nitrate reductase, in ubiquinol oxidation. Mol Microbiol 44: 245–255.
Brondijk THC, Nilavongse A, Filenko N, Richardson DJ, Cole JA . (2004). NapGH components of the periplasmic nitrate reductase of Escherichia coli K-12: location, topology and physiological roles in quinol oxidation and redox balancing. Biochem J 379: 47–55.
Cruz-Garcia C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM . (2007). Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol 189: 656–662.
Drummond AJ, Aston B, Buxton S, Cheung M, Heled J, Kearse M et al. (2010). Geneious v4.8. Available from http://www.geneious.com.
Edwards RA, Keller LH, Schifferli DM . (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207: 149–157.
Gao HC, Yang ZK, Barua S, Reed SB, Romine MF, Nealson KH et al. (2009). Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J 3: 966–976.
Gao HC, Yang ZK, Wu LY, Thompson DK, Zhou JZ . (2006). Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol 188: 4560–4569.
Gralnick JA, Vali H, Lies DP, Newman DK . (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. P Natl Acad Sci USA 103: 4669–4674.
Gross R, Eichler R, Simon J . (2005). Site-directed modifications indicate differences in axial haem c iron ligation between the related NrfH and NapC families of multihaem c-type cytochromes. Biochem J 390: 689–693.
Huelsenbeck JP, Ronquist F . (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755.
Jepson BJ, Marietou A, Mohan S, Cole JA, Butler CS, Richardson DJ . (2006). Evolution of the soluble nitrate reductase: defining the monomeric periplasmic nitrate reductase subgroup. Biochem Soc Trans 34: 122–126.
Kern M, Simon J . (2008). Characterization of the NapGH quinol dehydrogenase complex involved in Wolinella succinogenes nitrate respiration. Mol Microbiol 69: 1137–1152.
Lenski RE, Rose MR, Simpson SC, Tadler SC . (1991). Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Naturalist 138: 1315–1341.
Maillard J, Spronk CAEM, Buchanan G, Lyall V, Richardson DJ, Palmer T et al. (2007). Structural diversity in twin-arginine signal peptide-binding proteins. P Natl Acad Sci USA 104: 15641–15646.
McGinnis S, Madden TL . (2004). BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32: W20–W25.
Miller TL, Wolin MJ . (1974). A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl Microbiol 27: 985–987.
Moreno-Vivian C, Cabello P, Martinez-Luque M, Blasco R, Castillo F . (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol 181: 6573–6584.
Murphy JN, Saltikov CW . (2007). The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J Bacteriol 189: 2283–2290.
Myers JM, Myers CR . (2000). Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J Bacteriol 182: 67–75.
Pittman MS, Elvers KT, Lee L, Jones MA, Poole RK, Park SF et al. (2007). Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol Microbiol 63: 575–590.
Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA . (1999). Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem J 344: 77–84.
Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ . (2001). Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci 58: 165–178.
Sears HJ, Little PJ, Richardson DJ, Berks BC, Spiro S, Ferguson SJ . (1997). Identification of an assimilatory nitrate reductase in mutants of Paracoccus denitrificans GB17 deficient in nitrate respiration. Arch Microbiol 167: 61–66.
Simpson PJ, Richardson DJ, Codd R . (2010). The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiol 156: 302–312.
Spector MP, del Portillo FG, Bearson SMD, Mahmud A, Magut M, Finlay BB et al. (1999). The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvationinducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiol-Uk 145: 3035–3045.
Stewart V, Lu Y, Darwin AJ . (2002). Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J Bacteriol 184: 1314–1323.
Stolz JF, Basu P . (2002). Evolution of nitrate reductase: molecular and structural variations on a common function. Chembiochem 3: 198–206.
Tabata A, Yamamoto I, Matsuzaki M, Satoh T . (2005). Differential regulation of periplasmic nitrate reductase gene (napKEFDABC) expression between aerobiosis and anaerobiosis with nitrate in a denitrifying phototroph Rhodobacter sphaeroides f sp denitrificans. Arch Microbiol 184: 108–116.
Thompson JD, Plewniak F, Poch O . (1999). A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res 27: 2682–2690.
Wang F, Li Q, Xiao X . (2007). A novel filamentous phage from the deep-sea bacterium Shewanella piezotolerans WP3 is induced at low temperature. J Bacteriol 189: 7151–7153.
Wang F, Xiao X, Ou HY, Gai YB, Wang FP . (2009). Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J Bacteriol 191: 2574–2584.
Wang FP, Wang JB, Jian HH, Zhang B, Li SK, Wang F et al. (2008). Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One 3: e1937.
Xiao X, Wang P, Zeng X, Bartlett DH, Wang FP . (2007). Shewanella psychrophila sp. nov. and Shewanella piezotolerans sp. nov., isolated from west pacific deep-sea sediment. Int J Syst Evol Microbiol 57: 60–65.
Acknowledgements
This work has been partly supported by Natural Science Foundation of China (grant number 40625016, 40776095, 40876086), by the National High-Tech Program (2007AA091904), and by COMRA fund (DYXM-115-02-2-03).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, Y., Wang, F., Xu, J. et al. Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME J 5, 843–855 (2011). https://doi.org/10.1038/ismej.2010.182
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.182
Keywords
This article is cited by
-
Transcriptomic Analysis Reveals Common Adaptation Mechanisms Under Different Stresses for Moderately Piezophilic Bacteria
Microbial Ecology (2021)
-
Physiological and transcriptional approaches reveal connection between nitrogen and manganese cycles in Shewanella algae C6G3
Scientific Reports (2017)
-
Characterization of the relationship between polar and lateral flagellar structural genes in the deep-sea bacterium Shewanella piezotolerans WP3
Scientific Reports (2016)
-
Development of a genetic system for the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913
Microbial Cell Factories (2014)
-
Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm
Scientific Reports (2014)