Abstract
In spite of the importance of plasmids in bacterial adaptation, we have a poor understanding of their dynamics. It is not known if or how plasmids persist in and spread through (invade) a bacterial population when there is no selection for plasmid-encoded traits. Moreover, the differences in dynamics between spatially structured and mixed populations are poorly understood. Through a joint experimental/theoretical approach, we tested the hypothesis that self-transmissible IncP-1 plasmids can invade a bacterial population in the absence of selection when initially very rare, but only in spatially structured habitats and when nutrients are regularly replenished. Using protocols that differed in the degree of spatial structure and nutrient levels, the invasiveness of plasmid pB10 in Escherichia coli was monitored during at least 15 days, with an initial fraction of plasmid-bearing (p+) cells as low as 10−7. To further explore the mechanisms underlying plasmid dynamics, we developed a spatially explicit mathematical model. When cells were grown on filters and transferred to fresh medium daily, the p+ fraction increased to 13%, whereas almost complete invasion occurred when the population structure was disturbed daily. The plasmid was unable to invade in liquid. When carbon source levels were lower or not replenished, plasmid invasion was hampered. Simulations of the mathematical model closely matched the experimental results and produced estimates of the effects of alternative experimental parameters. This allowed us to isolate the likely mechanisms most responsible for the observations. In conclusion, spatial structure and nutrient availability can be key determinants in the invasiveness of plasmids.
Similar content being viewed by others
Introduction
Plasmids play an important role in the evolution and rapid adaptation of bacteria and archaea by spreading multidrug resistance and many other traits among distantly related hosts (Mazodier and Davies, 1991; Dröge et al., 1999; van Elsas and Bailey, 2002; Smets and Barkay, 2005; Sørensen et al., 2005; Thomas and Nielsen, 2005). In spite of their importance in bacterial adaptation, we currently have a poor understanding of the population dynamics of plasmids in the absence of selection for the traits they encode. The success of plasmids as gene shuttles depends on their ability to transfer to and maintain themselves in a variety of hosts. It is easy to understand maintenance of plasmids in populations that are experiencing positive selection for plasmid-encoded traits (hereafter referred to as ‘selection’). Although it is much less obvious if and how these plasmids persist in a bacterial community that has no apparent selection for the plasmid, plasmid persistence is known to depend on several factors: (1) the ability to replicate in multiple hosts; (2) mis-segregation of plasmid copies during cell division (segregational loss) and post-segregational killing potential; (3) the relative growth rates of plasmid-bearing hosts and their plasmid-free counterparts, a function of the cost of the plasmid to its host; and (4) the conjugative transfer rate of the plasmid (Stewart and Levin, 1977; Levin et al., 1979; Proctor, 1994; Bergstrom et al., 2000). Therefore, persistence of a self-transmissible plasmid requires successful horizontal transfer and, perhaps, occasional selection to counteract the effects of segregational loss and (periods of) plasmid cost.
A key component in the long-term survival of populations is the ability to ‘invade when rare’. Changing environmental or biotic factors can eventually drive a population to extinction or low density in a given region. Survival on a larger scale depends on the ability to either remain viable long enough for favourable conditions to return or to disperse and colonize new territory starting from low densities (MacArthur and Wilson, 1967). Similarly, for a plasmid to increase its frequency in a bacterial population or community in the absence of selection, it should be able to invade plasmid-free cells at a high rate, much like a parasite. In this study, we define this phenomenon as plasmid invasion, that is, the ability of a plasmid to significantly increase its proportion in the population (hereafter referred to as ‘plasmid frequency’) when it is initially rare. It has been postulated that plasmid invasion is unlikely to occur in the absence of selection (Levin and Lenski, 1983). This conclusion was based on estimates of parameters of models developed by Stewart and Levin approximately 30 years ago (Stewart and Levin, 1977; Levin et al., 1979), and on plasmid transfer rates experimentally observed in liquid cultures with F and F-like plasmids. Few studies have examined plasmid invasion, and only one demonstrated plasmid invasion when the plasmid-bearing fraction of the population was initially rare. Lundquist and Levin (1986) found that in their chemostat cultures, two naturally occurring plasmids were able to invade a population from an initial frequency of 10−4 to almost 1. Although these results were intriguing and suggest that some drug resistance plasmids might be able to spread and persist within pathogen populations in the absence of any antibiotics, for 20 years thereafter, no additional studies examined the conditions under which plasmids can invade a population by infectious transfer when initially very rare. Turner (2004) further investigated plasmid invasion as a function of recipient densities to address questions of trade-off between vertical and horizontal transfer, but the initial donor/recipient ratio was not as low (ca. 5 × 10−3). Moreover, these studies were done only in liquid systems. There is a great lack of information on plasmid dynamics in spatially structured bacterial populations, that is, bacteria that grow in aggregates or on surfaces as microcolonies and biofilms. Here and throughout the paper, spatial structure refers to all surface-associated dynamics and the resultant spatial heterogeneities in the population (but not the environment). Given that the majority of bacterial populations are spatially structured (Costerton et al., 1994), we need to take into account the architecture of microbial communities when studying the kinetics of gene flow and plasmid invasion.
Plasmids of the incompatibility group P-1 (IncP-1 plasmids) transfer very efficiently to a wide range of hosts and are abundant in several environments (Thomas and Smith, 1987; Top et al., 2000; Smalla et al., 2006, Schlüter et al., 2007). They are known to transfer at much higher rates on solid substrates than in mixed liquids. It has been postulated that mating aggregates, formed and held together in part by the short rigid pili of the IncP-1 plasmids, are too fragile for efficient conjugative transfer in stirred or shaken liquid cultures (Bradley et al., 1980). Recently, Bahl et al. (2007a, 2007b) used a wild-type and conjugation-deficient mutant IncP-1 plasmid to show that the conjugation rate of an IncP-1 plasmid in bacteria growing on surfaces (filter on agar and the rat intestine) can be high enough to permit long-term persistence and even a 10-fold increase in plasmid frequency (from 0.1 to 1) in the absence of selection. This confirmed a previous report that conjugative plasmid transfer can play a key role in plasmid stability (Sia et al., 1995), but the ability of the plasmid to invade a population from an initial frequency lower than 0.1 was not examined.
The dynamics of spatially structured bacterial populations can be very different from those grown in mixed liquids and cannot be simulated by the ordinary differential equation models that assume complete mixing (Dieckmann et al., 2000). Conjugative plasmid transfer is no exception to this, as it requires cell contact and is therefore determined by the spatial configuration of a population or community. The failure of ordinary differential equation models to capture experimentally observed dynamics of plasmid transfer has already been demonstrated in several studies (Simonsen, 1990; Licht et al., 1999; Pinedo and Smets, 2005; Krone et al., 2007). Moreover, plasmids such as those of the IncP-1 group transfer at higher rates on solid substrates than in liquids. For these various reasons, the conclusions about plasmid invasiveness based on mass–action differential equation models and chemostat cultures with F-like and other plasmids as model systems cannot be simply extended to IncP-1 plasmids in spatially structured populations.
Among the empirical results concerning plasmid spread in spatially structured populations, some of the most intriguing ones have shown limitations to plasmid invasion. For example, the TOL plasmid pWWO transferred to cells at the edge of a colony of plasmid-free cells, but could not invade further into the colony in the absence of selection (Christensen et al., 1996; Häagensen et al., 2002). A similar observation was made for bacteria grown in biofilm flow cells, where the deeper layers of the biofilm were not invaded by the plasmid (Christensen et al., 1998). Hypotheses have been raised about possible plasmid transfer inhibition mechanisms, or the lack of nutrient availability in those deeper cell layers. Nutrients have been shown to positively affect plasmid transfer efficiency in some cases, but not in others (Hausner and Wuertz, 1999; van Elsas and Bailey, 2002), and therefore it remains unclear why a plasmid does not completely invade a colony or biofilm, and if specific conditions may be conducive to plasmid invasion (Licht et al., 1999; Molin and Tolker-Nielsen, 2003).
The goal of this study was to use a joint experimental/theoretical approach to try to understand the factors that promote plasmid invasion in a bacterial population when the initial plasmid frequency is extremely low (as low as 10−7). Specifically, we hypothesized that self-transmissible IncP-1 plasmids can invade a population of plasmid-free hosts in the absence of selection when initially very rare, but only in spatially structured habitats and when nutrients are regularly replenished. The model plasmid was the multidrug resistance plasmid pB10, a 64.5 kb IncP-1β plasmid that was isolated from a wastewater treatment plant in Germany (Dröge et al., 2000; Schlüter et al., 2003). Both experimental and simulation results showed that, under certain conditions of spatial cell organization and nutrient availability, plasmid pB10 was able to invade Escherichia coli hosts and almost completely infect the population, increasing its frequency from 10−7 to almost 1.
Materials and methods
Media and culture conditions
Luria broth (LB) medium, LB agar (LBA) medium and mineral salts medium M9 were prepared according to Sambrook and Russell (2001), except that M9 was supplemented with 5 mg l−1 of trace elements (Stanier et al., 1966). All plasmid transfer, invasion, stability and fitness experiments were performed at 37 °C using M9 medium with 2 g l−1 glucose and 15 g l−1 agar unless otherwise stated. Antibiotics were used at the following concentrations unless otherwise mentioned: 50 mg l−1 rifampicin (Rif), 30 mg l−1 nalidixic acid (Nal) and 10 mg l−1 tetracycline (Tc). Antibiotic concentrations used in the media are abbreviated according to the following pattern: LBRif50 stands for LB medium with Rif (50 mg l−1).
Strain construction
The bacterial strain used in this study was E. coli K12 MG1655, henceforth briefly referred to as K12. To enable selective plating, Rif-resistant (RifR) and Nal-resistant (NalR) mutants were obtained as described previously (De Gelder et al., 2007). The RifR mutant (K12Rif) was typically used as the plasmid donor (initial plasmid-bearing cells), and the NalR mutant (K12Nal) as recipient (initial plasmid-free cells). To examine the possibility of bias in this choice of donor and recipient, a few experiments were performed with K12Nal serving as the donor and K12Rif as recipient. Recipient cells that received the plasmid are referred to as transconjugants.
The plasmid used in this study is the IncP-1β plasmid pB10 (Dröge et al., 2000; Schlüter et al., 2003), which confers resistance to the antibiotics Tc, streptomycin, amoxicillin and sulphonamides, and to HgCl2. This plasmid was transferred to K12Rif by conjugation on LBA using E. coli DH5α(pB10) as the donor (Lejeune et al., 1983), followed by selection for K12Rif (pB10) on LBA with Rif (50 mg l−1) and Tc (10 mg l−1).
Strains E. coli K12Nal::gfp and E. coli K12Rif(pB10::rfp) were constructed by insertion of a mini-transposon containing a kanamycin resistance gene and a gene coding for the green fluorescence protein (gfp) or red fluorescence protein (dsRed, also called rfp), using vectors pJBA120 and pSM1833, respectively (Häagensen et al., 2002; De Gelder et al., 2005). The location of the rfp cassette insertion in pB10 was determined as described previously (De Gelder et al., 2005).
Tests for plasmid stability, cost and transferability
Stability experiments were performed in liquid medium as described previously (De Gelder et al., 2007). Briefly, after growing precultures with Tc, bacterial cultures were grown in the absence of antibiotics for approximately 100 generations (approximately 10 generations per day). At regular time points, cultures were plated onto LB and 50 colonies were replicated onto LBTc and LB, and the fraction of TcS, and thus plasmid-free cells, was calculated.
Pairwise competition experiments were performed to determine the relative fitness of different strains and the cost of plasmids pB10 and pB10::rfp. To determine plasmid cost, plasmid-free strains were competed against their plasmid-bearing counterparts (either pB10 or pB10::rfp). Competition experiments were performed as described previously (Bouma and Lenski, 1988; Sota et al., 2007) and the plasmid cost (c) was calculated as 1−W, where W is the relative fitness (Heuer et al., 2007).
To verify that genetic changes in strains or plasmids, due to the insertion of fluorescent protein genes or selection for resistance mutations, did not affect plasmid transfer frequencies, filter matings were performed with all strain derivatives used in the invasion experiments. Cultures of plasmid-bearing donor cells and plasmid-free recipient cells, grown overnight in the presence of appropriate antibiotics (for example, in LBRif50Tc10 for K12Rif(pB10) and in LBNal30 for K12Nal), were centrifuged, decanted and resuspended in the same volume. A 5 μl droplet of recipient cells was placed in the centre of a 25 mm polycarbonate filter (Isopore membrane filter 0.4 μm HTBP; Millipore, Danvers, MA, USA) on top of M9 agar. As soon as the droplet was absorbed into the filter, a 5 μl droplet of donor cells was placed directly on the recipient cells and left to dry. The remaining filters were incubated for 2 h at 37 °C, and subsequently transferred to 2 ml of saline. The cells were suspended using a vortex mixer, diluted and plated at appropriate antibiotic concentrations to enumerate the number of donor, recipient and transconjugant cells. The ‘transfer frequency’ after the 2 h incubation period was calculated by dividing the density of transconjugant cells by the density of donor cells.
Plasmid invasion experiments
Five ‘plasmid invasion’ protocols were designed to determine the extent of plasmid spread through a population of plasmid-free bacteria in the absence of antibiotics, when the initial donor fraction was very low (10−7). The protocols described in detail below were performed in triplicate and were initiated as follows. Cells of donor and recipient strains were harvested from 1 ml overnight LB cultures (grown in the presence of appropriate antibiotics) by centrifuging, decanting the supernatant, resuspending the cells in saline, and centrifuging and decanting again. The recipient cells were resuspended in 100 μl of saline and the donors in 1 ml, and the donor cell suspension was further diluted to 10−4. For invasion experiments performed on surfaces, a 100 μl aliquot of recipient cells and 1 μl of the 10−4 donor cell suspension were mixed and immediately spread plated (using a disposable cell spreader) on a 47-mm diameter polycarbonate filter (Isopore membrane filter 0.4 μm HTBP, Millipore), which was placed on top of M9 glucose agar (∼5 ml) in a 60 mm petri dish. This resulted in approximately 5 × 107 recipient cells per square centimetre and approximately 5 donor cells per square centimetre, and therefore an initial donor/recipient ratio of 10−7. For the invasion experiment performed in liquid medium, the same procedure was followed to initiate the experiments except that the donor and recipient cell mixture was transferred to 2 ml liquid M9 glucose medium in 15 ml disposable conical tubes (Nalge Nunc International, Rochester, NY, USA). These cultures were incubated on a rotary shaker (200 r.p.m.).
As filters (or tubes in the case of experiments in liquid medium) had to be sacrificed for enumeration of the donor, recipient and transconjugant populations at each time point, triplicate filters and tubes for each time point were set up at day 0. The following protocols were used, after initiating the experiments, to test the effects of spatial structure and nutrient availability on plasmid invasion:
Protocol 1 (undisturbed filter transfer). Bacteria were grown on filters and the filters were transferred daily to fresh agar medium by using flame-sterilized forceps. This protocol provided the cells daily with fresh M9 glucose medium without altering the spatial structure of the population.
Protocol 2 (disturbed filter transfer). The spatial structure of the bacterial population was disrupted just before each daily transfer by removing the bacteria from the filters and suspending them in 2 ml saline through 1 min of vortex mixing. The suspension was then concentrated by centrifugation and resuspension of the pellet in 100 μl saline, and transferred to a new filter on a new agar plate by spread plating. Cells were thus provided with fresh medium everyday, as in protocol 1, but they were also redistributed over the filter in such a way that their contact with neighbouring cells and proximity to the agar medium or air/colony interface was randomly changed.
Protocol 3 (no filter transfer). The filter remained on the same agar plate for the entire period of the 15-day long experiment. Thus, the initial supply of nutrients was not replenished and the spatial structure was maintained throughout the experiment.
Protocol 4 (liquid medium transfer). The disposable conical tubes were centrifuged daily, the supernatant was decanted and the cells were resuspended in 2 ml of fresh medium. Thus, cells were provided daily with fresh M9 glucose medium, as in protocols 1 and 2, but there was no spatial structure as the cultures were continuously mixed in liquid medium on a rotary shaker at 200 r.p.m.
Protocol 5 (undisturbed filter transfer-glucose limitation). This was similar to protocol 1, but used M9 medium augmented with varying levels of glucose (2, 0.2, 0.02 and 0 g l−1).
Once the protocols were set up, the donor, recipient and transconjugant cell populations were enumerated as follows: on day 0 and at various other time points, cells were resuspended in 2 ml saline (as described under protocol 2), and appropriate dilutions (in saline) were plated on LBA plates containing appropriate antibiotic concentrations; these concentrations typically were LBRif50Tc10, LBNal30 and LBNal30Tc10, respectively. This was done using a Spiral Plater Autoplate 4000 and a Q-Count colony counting system (Spiral Biotech, Norwood, MA, USA). In all figures, the number of recipient cells represents the number of plasmid-free, rather than total, recipients, and was calculated as the difference between cell counts on LBNal30 (representing all recipients including transconjugants) and those on LBNal30Tc10 (representing transconjugants). In one experiment (Figure 1b), after day 5, the number of plasmid-free recipients could no longer be determined by subtracting the number of transconjugants from the total number of recipients, as they were indistinguishable within experimental error. Therefore, the fraction of plasmid-free clones within the recipient population was determined via replica plating by transferring colonies for each of the triplicate experiments from LBANal30 onto LBATc10, and calculating the fraction of Tc-sensitive clones.
Effect of spatial structure and nutrients on plasmid invasion. The dynamics of E. coli K12 donors (dotted, •), recipients (dashed, ▪) and transconjugants (solid, ▴) on filters on M9 agar plates with 2 g l−1 glucose are shown. (a–c) Experimental data; (d–f) simulations. Invasion experiments were initiated at a donor/recipient ratio of 10−7 and three protocols were followed. (a and d) the spatial structure was left intact with daily transfer of the filters; (b and e) the spatial structure was disturbed with each daily transfer of the filters; (c and f) the spatial structure was left intact and there was no daily transfer of the filters, and thus no nutrient replenishment. Data present averages and s.d. of triplicate experiments and simulations. Grid size for simulation: 2000 × 2000; parameters: ΨR=0.5 h−1, ΨT=0.48 h−1, ΨD=0.38 h−1, γ=2 h−1, L=9 h, m=16 cells, τ=0, pg=0.5 and pc=0.9.
Microscopy
To analyse the spatial patterns of the spread of plasmid pB10 by stereomicroscopy, similar ‘invasion experiments’ were performed as described above, using K12Rif (pB10::rfp) as the donor and K12Nal::gfp as the recipient strain. In this way, the donor bacteria were represented by red, the recipients by green and the transconjugants could be distinguished by the overlap of red and green signals. To enable detection of donor cells, the donor/recipient ratio was 1000 × higher than in the other invasion experiments, that is, 10−4. At the time of inoculation (day 0), and on days 2 and 3, plates were viewed with a fluorescence stereoscope (LeicaMZ16F, Leica, Bannockburn, IL, USA) at × 115 magnification and images were taken with a digital camera (MicroPublisher 3.3 RTV, Qimaging, Surrey, British Columbia, Canada). Images were captured using the Qcapture Suite (QImaging), and then further processed for display using ImageJ (NIH).
Statistical analyses
The standard Student's t-test was used to determine the significance of invasion by comparing fractions of plasmid-bearing cells on days 0 and 15, and to determine whether final plasmid-bearing fractions were significantly different between the various plasmid invasion protocols. The t-test was also used to determine significant differences in plasmid transfer frequencies between marked and unmarked plasmids. In each of these analyses, a significance level of 0.05 was used.
Mathematical model
Before describing the mathematical model used to simulate plasmid dynamics on surfaces, we outline the features of the experimental system we wish to capture. When cells have grown to a significant density on the plate (by the end of day 1), nutrients, diffusing from the agar through the filter into the cell layer (here called a colony), are limited due to consumption by cells; thus, nutrients are plentiful only at the beginning of a passage, before they are depleted, and diminish in concentration as one moves up the colony away from the nutrient source. This differential access to nutrients is a consequence of the three-dimensional (3D) structure of the colony and leads to a nutrient concentration gradient, with higher concentration at the bottom than at the top. One of our hypotheses is that nutrient levels influence plasmid spread. To address this while keeping the model simple, we distinguish only between cells that have access to nutrients and cells that do not, instead of modelling the full time-dependant 3D gradient of nutrients. Although this is an obvious oversimplification, we claim it is sufficient to capture the main effects of nutrients and spatial structure in the experiments, and match the empirical observations. That such a simplification of the underlying mechanisms of nutrient diffusion would be enough to accurately simulate the multiple features of the experimental results suggests that these mechanisms play a dominant role in determining those features. Indeed, we show below that the qualitative behaviour of the simulations is quite robust to changes in model parameters. This further suggests, not surprisingly (yet nontrivially), that more complex 3D biofilm models with more detailed modelling of nutrient diffusion should predict the same results.
The mathematical model is built on a 2D (rather than 3D) square lattice of sites, but with certain essential aspects of the 3D structure codified by distinguishing between two ‘levels’ of cells (‘top’ and ‘bottom’) and designating a time interval [0, L] just after each transfer of the population to fresh medium during which nutrients are available to all bottom cells. The labels ‘top’ and ‘bottom’ are used to suggest that the cells in the bottom category have access to nutrients diffusing up from the agar, whereas those in the top category do not. We assign these labels in such a way that cells in the ‘top’ category can only be assigned when all the ‘bottom’ slots are filled. The maximum number of cells at a site of the lattice is assumed to be M. The first m of these (m⩽M) to appear are considered ‘bottom’ cells, and have access to nutrients (when present); the remaining cells (up to M−m of them) appearing at the site are considered ‘top’ cells and cannot access nutrients. Cells that have access to nutrients are said to be ‘active’, meaning that they are capable of reproduction and conjugation; cells without access to nutrients are called ‘inactive’ and cannot take part in reproduction or conjugation.
Our model is from the class of interacting particle systems or stochastic cellular automata. Because of the fact that sites on the lattice are updated using only summary statistics that condense the 3D information about the colony, this model can be thought of as a stochastic ‘coupled map lattice’ approximation to an individual-based model. A related interacting particle system model of plasmid population dynamics was developed by us earlier (Krone et al., 2007), but that model did not include the aspects of 3D structure that are essential in this study. Simulations of the model were carried out on a 2000 × 2000 lattice with periodic boundary; the commonly used periodic boundary condition, equating the top edge of the lattice to the bottom edge and the left edge to the right edge, eliminates complicating edge effects. Each site of the lattice can contain cells of several types: donors (D), recipients (R) and transconjugants (T). In addition to these types, each cell also carries a designation of top or bottom that does not change in the ‘undisturbed filter transfer’ simulations, but can change in the ‘disturbed filter transfer’ simulations.
A ‘local neighbourhood’ specifies which sites are neighbours for the purposes of determining conjugation events and the locations of daughter cells after cell division. We took the local neighbourhood of a given site ‘x’ to be the nine sites on the lattice consisting of the eight adjacent sites plus the site itself (that is, all the sites in the 3 × 3 box centred at x). At each step of a simulation, a ‘focal site’ was randomly chosen and, by comparing the rates of various events, it was determined whether a change should be made to the focal site (or one of its neighbours) and, if so, what that change would be. Thus, the focal site can be thought of as initiating a local change in the current configuration. We synchronized the time scale in the simulations with actual time in the experiments by setting the doubling time of recipient cells at low density to 80 min. This doubling time was determined experimentally by monitoring recipient cell growth every hour during the first 8 h of a typical invasion experiment on M9 glucose (2 g l−1) agar (data not shown).
Rates (of reproduction and conjugation) in the model depend on nutrient levels only indirectly through the parameters m and L that can be roughly thought of as proxies for nutrient diffusivity and amount of daily nutrient, respectively. We specified constant growth rates ΨR, ΨD and ΨT for R, D and T, respectively, and a conjugation rate γ. These rates were assumed to be in effect when the cells are ‘active,’ and otherwise the corresponding rates were 0. When an active cell at the focal site is chosen to reproduce, the offspring is placed at the same site as the ‘parent’ with probability 1−pg, and at one of the eight neighbouring sites with probability pg (the latter probability is sometimes called a ‘coupling parameter’ in the literature on coupled map lattices). Similarly, 1−pc was used for the probability that a conjugation event initiated by a plasmid-bearing cell at the focal site occurs with a plasmid-free cell at the same site. Segregative plasmid loss occurs with probability τ when a daughter of a plasmid-bearing cell (D or T) fails to receive a plasmid molecule during cell doubling. Thus, when a transconjugant divides, the daughter cell will be a T with probability 1−τ (resulting in vertical transmission of the plasmid) and an R with probability τ. Let  be the number of R's at the focal site (w for ‘within’) and
be the number of R's at the focal site (w for ‘within’) and  the total number of R's at the eight neighbouring sites (nbr for ‘neighbour’), with similar designations for D and T. The number of active cells in each category is denoted using a subscript ‘act’; for example,
the total number of R's at the eight neighbouring sites (nbr for ‘neighbour’), with similar designations for D and T. The number of active cells in each category is denoted using a subscript ‘act’; for example,  indicates the total number of R's with access to nutrients at the eight sites neighbouring the focal site. Furthermore, we write
indicates the total number of R's with access to nutrients at the eight sites neighbouring the focal site. Furthermore, we write  for the amount of vacant ‘space’ at the focal site and
for the amount of vacant ‘space’ at the focal site and  the total amount of vacant space at the eight neighbouring sites. If we divide the ‘within’ quantities by M, we get the fraction of the focal site consisting of that state:
the total amount of vacant space at the eight neighbouring sites. If we divide the ‘within’ quantities by M, we get the fraction of the focal site consisting of that state:  , where the • can represent any of the subscripts. Similarly, dividing the ‘neighbour’ quantities by 8M yields the corresponding fractions:
, where the • can represent any of the subscripts. Similarly, dividing the ‘neighbour’ quantities by 8M yields the corresponding fractions:  . The rate at which the focal site produces a new R is
. The rate at which the focal site produces a new R is

whereas new D's appear at rate

and new T's appear at rate

In the last expression, terms involving γ correspond to conjugation events and, in addition to an increase in the number of T's at the site of the recipient selected for conjugation, there is the corresponding loss of an R at that site. In a reproduction event, when an offspring is placed at a neighbouring site, corresponding to terms involving the factor  , the site is chosen at random from among the eight neighbours, with each site's probability proportional to the fraction of vacant space at that site. In all cases, the new cell will be in the bottom level unless there are already at least m cells at that site. Note that, a site holding a full complement of M cells cannot accept any offspring. The per capita rate of growth for recipients,
, the site is chosen at random from among the eight neighbours, with each site's probability proportional to the fraction of vacant space at that site. In all cases, the new cell will be in the bottom level unless there are already at least m cells at that site. Note that, a site holding a full complement of M cells cannot accept any offspring. The per capita rate of growth for recipients,  , is expressed in a way that depends on the number of vacant sites (and hence cell density) at the focal and nearby sites. There are similar expressions for T and D. Note that, as the number of vacant sites decreases due to an increase in local population density, the effective growth rate decreases. Thus, the combination of a nutrient clock (L) and density-dependant growth accounts for the fact that, as population density increases, a larger proportion of the nutrient goes toward cell maintenance. An overview of all model parameters is given in Table 1.
, is expressed in a way that depends on the number of vacant sites (and hence cell density) at the focal and nearby sites. There are similar expressions for T and D. Note that, as the number of vacant sites decreases due to an increase in local population density, the effective growth rate decreases. Thus, the combination of a nutrient clock (L) and density-dependant growth accounts for the fact that, as population density increases, a larger proportion of the nutrient goes toward cell maintenance. An overview of all model parameters is given in Table 1.
Finally, to keep the model description simple, we have omitted from the model plasmid-free cells that arise through segregative loss in donor cells. These cells are of a different type than the recipient cells, but have no noticeable effect on the simulations in this study (data not shown), as donor frequencies always remained low.
In the simulations of the ‘undisturbed filter transfer’ protocol, we simply reset the nutrient clock to 0 at the start of each passage (so that there are L h of nutrient accessibility for cells in the active zone), leaving the spatial structure and cell states unchanged. In simulations of the ‘disturbed filter transfer’ protocol, the start of a new passage involved randomly distributing the cells among the sites with at most M per site, and then randomly determining which cells should go in the active zone; any cells remaining after filling the active zone are put in the inactive zone.
Note that, unlike the mass–action models that are based on bulk concentrations and densities that must be specified in terms of units, the individual-based nature of the interacting particle system model means that ‘rates’ simply signify (inverse) times until the specified events occur. To see how length scales in the interacting particle system model compare to those in experiments, each lattice site should be thought of as the location of a microcluster of up to M cells. Thus, accounting for a cell size of about 1 or 2 μm and intercellular distances also in the order of micrometres, and with 2000 lattice points per side for a typical simulation grid in this paper, we should consider our ‘simulation window’ to be one to several millimetres per side.
Results
Effect of spatial structure on plasmid invasion
The effect of spatial structure on the ability of plasmid pB10 to invade a plasmid-free population of bacteria when initially rare was tested under a variety of conditions. We defined invasion as a significant increase in the fraction of plasmid-bearing cells from day 0 to 15 (P=0.05) in the absence of selection for the plasmid. Three plasmid invasion experiment protocols were compared, ‘undisturbed filter transfer’ (Figure 1a), ‘disturbed filter transfer’ (Figure 1b) and ‘liquid medium’ (Figure 2), each providing daily nutrient replenishment. First, there was a stark contrast between the significant plasmid invasion (P<0.001) on agar under the undisturbed filter transfer protocol (Figure 1a) and the inability of the plasmid to invade in a mixed liquid environment (Figure 2). Although the T densities increased sharply in these spatially structured populations (Figure 1a), the D and R densities remained relatively steady. This strongly suggests that the observed plasmid invasion was caused by horizontal transfer from donor and especially transconjugant cells to the dense lawn of recipients. Second, comparison of the results of the disturbed and undisturbed filter transfer protocols (Figures 1b and a, respectively) shows that combinations of 24 h periods of sustained spatial structure with daily disturbance of that structure were more conducive to plasmid invasion. Under this protocol, the transconjugants became the numerically dominant population, and were 50 times more abundant than plasmid-free recipient cells by day 15 (Figure 1b). Thus, daily nutrient replenishment, in combination with rearrangement of the spatial structure, resulted in an almost complete invasion of the E. coli population by plasmid pB10.
Dynamics of E. coli K12 donors (dotted, •), recipients (dashed, ▪) and transconjugants (solid, ▴) in mixed liquid M9 medium with 2 g l−1 glucose. Invasion experiments were initiated at a donor/recipient ratio of 10−7 and the liquid medium was exchanged daily. Data present averages and s.d. of triplicate experiments.
Parameter estimation in the mathematical model
The parameters in the mathematical model (Table 1) were chosen to match multiple features of the experimental data. It is important to note that we required the same parameters to be used in both the ‘undisturbed filter transfer’ and ‘disturbed filter transfer’ models. In matching the population densities of R, D and T over time in each experiment, we paid special attention to the initial rates of rapid increase (with R and D depending on cell growth, and T depending on both growth and conjugation) before the stationary phase, and the stationary phase densities, including the final difference between T and R densities.
The growth rates ΨR, ΨD and ΨT were estimated first, using independent filter experiments, as these rates were independent of the other parameters. For example, to match the doubling time of 80 min=1.33 h for recipients growing under conditions similar to those of the invasion experiments (data not shown), we set the per capita growth rate at ΨR=0.5 (as e(0.5)(1.33)≈2) when there were four cells per site. Once these rates were set in the model, we began choosing the parameters L, m and γ. The procedure involved systematically eliminating parameter combinations that were not consistent with the observed time-course data (Figure 1). The allowable values of m and L were quickly restricted by matching the eightfold increase in population size that was observed on the first day of the experiment (Figure 1). For each such pair of values, the conjugation rate was varied and simulations were compared with the trajectories of the transconjugant densities (in both disturbed and undisturbed protocols) (Figure 3). This led to a very narrow range of parameters that were consistent with the empirical data. Additional simulations showed that the results were very insensitive to the value of τ, as well as to our choice of coupling parameters; a large change (for example, increasing pg from 0.5 to 0.9), however, would require an adjustment of the other parameters. An important feature captured by the model is the large difference in the extent of plasmid invasion between undisturbed and disturbed filter transfer experiments. Although the model is an abstraction of the full biological system, it appeared to capture the most important mechanisms that determine the rates and extent of plasmid invasion under the different protocols. The final choice of parameters for fitting simulations to empirical data (Figure 1) was made by minimizing the mean squared error over all time points and experimental conditions; parameter values are summarized in Table 1.
Effect of parameters L and m on simulations of plasmid invasion. The dynamics of E. coli K12 donors (dotted, •), recipients (dashed, ▪) and transconjugants (solid, ▴) with different values of L and m are shown. Other parameters: ΨR=0.5 h−1, ΨT=0.48 h−1, ΨD=0.38 h−1, γ=2 h−1, τ=0, pg=0.5 and pc=0.9; grid size for simulation: 2000 × 2000.
Effect of nutrient concentration on plasmid invasion
To examine if nutrient concentration is critical for successful plasmid invasion in an E. coli population, we compared results from the ‘undisturbed filter transfer’ protocol (Figure 1a), described above, with those of the ‘no filter transfer’ protocol (Figure 1c). As shown in Figure 1c, when nutrients were not replenished daily, initial horizontal plasmid transfer occurred, but there was no further increase in the number of transconjugants after day 1. This resulted in a much lower final fraction of plasmid-bearing cells than when nutrients were replenished daily (3.0 × 10−5 versus 1.3 × 10−1; P<0.001). A similar behaviour was observed in simulations of the model with no filter transfer (Figure 1f), where once the original nutrients were depleted, all densities remained constant. One feature in the experiments that was not present in the model was the slight decay of cell densities under the ‘no filter transfer’ protocol. This was most likely a result of cell death, which was not included in the model. Our findings strongly suggest that prolonged availability of nutrients is critical for an IncP-1 plasmid to continue to spread through an E. coli population in the absence of any known selection for the plasmid.
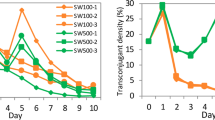
To further examine the effect of nutrients on the ability of the plasmid to invade an E. coli population, ‘undisturbed filter transfer’ experiments were carried out at several concentrations of glucose (2, 0.2, 0.02 and 0 g l−1). There was no significant difference in the final plasmid-bearing cell fractions between the experiments with the two highest glucose concentrations after 15 days, whereas when the concentration was only 0.02 g l−1, the fraction was significantly lower on day 15 (P=0.002) (Figure 4). However, even at this low glucose concentration, the plasmid-bearing fraction further increased when we continued the experiment, and by day 38 it was significantly higher than the initial fraction (P<0.001). When no glucose was added to the medium, transconjugants were formed, but their density stabilized at a much lower level, and no significant invasion was observed (Figure 4). Thus, plasmid invasion was positively correlated with the amount of carbon source in the medium, with the greatest sensitivity occurring at very low concentrations.
Effect of different glucose concentrations on plasmid invasion. The dynamics of E. coli K12 donors (dotted, •), recipients (dashed, ▪) and transconjugants (solid, ▴) on filters on M9 agar plates with different concentrations of glucose or no glucose are shown. Invasion experiments were initiated with a donor/recipient ratio of 10−7. Data present averages and s.d. of triplicate experiments.
The simulations of the mathematical model also captured the dependence of plasmid invasion on nutrient availability (Figure 3). This was performed by varying the values of L and m. Of course, as these parameters are abstractions meant to mimic aspects of nutrient level and diffusivity, we were not able to set up an exact correspondence between glucose levels and values of L and m. Nevertheless, these simulations further validate the hypothesis that access to nutrients and spatial distribution control the rate and extent of plasmid invasion.
Microscopy of plasmid invasion
To visualize the process of plasmid invasion, an invasion experiment was set up with an initial D/R ratio of 10−4 under the undisturbed filter transfer and disturbed filter transfer protocols. The filters were observed microscopically and micrographs were taken at three time points (Figure 5). After 3 days of the disturbed filter transfer protocol, the recipient population was completely transformed into transconjugants (Figure 5b), whereas the recipient population experienced visibly less invasion in the undisturbed filter transfer protocol (Figure 5a). These visual observations qualitatively confirm the findings presented above, obtained by cell enumeration, that spatial organization mediates the extent of plasmid invasion.
Stereoscope photographs of invasion experiments with an initial donor/recipient ratio of 10−4, at a magnification of × 115. Two protocols were followed: (a) the spatial structure was preserved during daily transfer of the filter and (b) the spatial structure was disturbed daily during transfer of the filter. Recipient cells are green; the entire field of each frame has a recipient-covered background. Donor cells are red and transconjugants are yellow/orange.
Effects of Rif resistance and Nal resistance mutations on plasmid invasiveness
To determine the effects of the Rif and Nal resistance mutations in hosts K12Rif and K12Nal on the growth rate (fitness) of the host and the plasmid transferability, relative fitness values and transfer frequencies were determined. There was no significant difference in the transfer frequency (T/D) between matings with K12Rif as donor and K12Nal as recipient, and matings with K12Nal as the donor and K12Rif as the recipient (2.36 × 10−1 and 2.71 × 10−1, respectively). From results of pairwise competition experiments in liquid medium, the relative fitness of K12Rif versus K12Nal was 80%, indicating that the RifR mutation has a large negative effect on the host fitness.
To determine if this growth rate difference between K12Rif and K12Nal in liquid medium had an effect on plasmid invasion, we reversed the strains used in the ‘undisturbed filter transfer’ invasion experiment. Thus K12Nal was now the donor and K12Rif the recipient strain. A similar drastic increase in transconjugant density was observed over 15 days, resulting in significant invasion (P<0.001) (data not shown). Thus, even in the case where the transconjugant cells (K12Rif(pB10)) grew more slowly than the donor cells (K12Nal(pB10)), the transconjugant fraction significantly increased (P<0.001) over time in the presence of nutrients, whereas the donor fraction did not. Given that the plasmid cost was approximately 5% (see below), these results confirm that the rise in transconjugants, and thus plasmid invasion, can only be explained by continued horizontal transfer of pB10 into the numerically abundant recipient cells.
Effects of rfp gene insertion on plasmid pB10 transfer, stability and cost
The stability, cost and transferability of pB10 and pB10::rfp were compared to verify if results from the microscopic analysis, performed with pB10::rfp, could be compared with all other invasion experiments conducted with the wild-type plasmid pB10. Our assays did not detect an effect of the insertion of the miniTn5-rfp transposon on these plasmid characteristics (data not shown). Sequence determination showed that the minitransposon was inserted into the klcB gene, a part of the klc/kle operon. Although the operon is known to have a role in plasmid stability, the function of klcB is not known (Adamczyk and Jagura-Burdzy, 2003).
Discussion
Our study has shown that the IncP-1β broad-host-range multidrug resistance plasmid pB10 can invade a plasmid-free population in the absence of antibiotics under certain conditions. Although pB10 donor cells were introduced at an extremely low density of approximately 5 c.f.u. cm−2 in a population of 107–108 plasmid-free recipient cells, invasion occurred, and after 2 weeks the plasmid-bearing cells represented up to 98% of the population, that is, 109 c.f.u. cm−2, the carrying capacity of the system. Our results suggest that either (1) IncP-1 plasmids can act as genetic parasites due to their high infectious transfer rates or (2) unknown selection for plasmid pB10 existed in a spatially structured environment. For example, Ghigo (2001) showed that IncP-1 and other plasmids promote biofilm formation. However, a similar form of improved adhesion to the filter cannot have been under selection in our experimental system because cells were retained on the filter, that is, there was no cell washout. Thus, the data strongly suggest that in E. coli populations grown on surfaces, plasmid pB10 can act as a genetic parasite due to its high infectious transfer rate.
Microscopic observations of plasmid transfer in bacterial colonies on agar media and in biofilms (Häagensen et al., 2002; Molin and Tolker-Nielsen, 2003) have shown that plasmid transfer occurred only at the interface of plasmid-bearing and plasmid-free cells, and the wave of infectious transfer did not extend very far into the colony or biofilm. As agar plates have a limited nutrient supply and steep nutrient gradients exist in biofilm flow cells, we hypothesized that nutrient limitation explains the previously observed lack of extensive plasmid invasion in a colony or biofilm. Our results support this hypothesis, as plasmid invasion was more pronounced with increasing concentrations of glucose (Figure 4) and when nutrients were replenished daily (Figure 1a compared with c). Thus drug resistance plasmids may be able, at least temporarily, to spread through a bacterial population in the absence of antibiotics as long as sufficient nutrients are available.
So far as we know, the only other study that examined plasmid invasion in a bacterial population from a very low fraction of plasmid-bearing cells was the one by Lundquist and Levin (1986). However, their study was performed in liquid medium in completely mixed chemostat reactors, with plasmids other than those of the IncP-1 group. They concluded that two of the seven plasmids they studied were able to invade because of their very high transfer rate during the period of transitory derepression of transfer, right after a plasmid enters a recipient cell, and that the invasion was dependant on the high cell density of the recipient population. IncP-1 plasmids like pB10 are known to transfer very efficiently on surfaces (Bradley et al., 1980). Thus, our data support the authors’ conclusion that under some conditions, plasmids can be parasitic owing to their high conjugative transfer rate. One factor that the authors did not mention in 1986 was the continuous supply of nutrients to the bacterial populations, a characteristic of chemostats. Moreover, no systematic comparison has ever been made between plasmid invasiveness in liquids (no spatial structure) and on surfaces (spatial structure). This is surprising, given the known differences in transfer efficiencies under these two conditions for different plasmid types (Bradley et al, 1980), and the important effects of spatial structure on the fate of plasmids (Simonsen, 1990; Krone et al., 2007).
Our finding that plasmid pB10 can infectiously spread through an E. coli population supports the results by Bahl et al. (2007a, 2007b), who observed that the IncP-1 plasmid pMIB4 was able to spread through a bacterial population on agar and in the rat gastrointestinal tract. On agar, horizontal transfer of plasmid pMIB4, which was initially present in 10–90 of the population, resulted in 100% plasmid-bearing cells after 3 days. By including tests with a transfer-deficient plasmid variant, they also showed that horizontal transfer, and not differential fitness, caused this increase in the fraction of plasmid-bearing cells on agar and counteracted plasmid loss in the rat intestine. Although we did not use a transfer-deficient mutant of pB10 in our study, the observed drastic increase (a factor 106–107) in the fraction of transconjugants in both the E coli K12Nal and K12Rif populations can only be explained by infectious plasmid spread. Our work also differed from their work in two important ways. The first difference was in the degree of plasmid invasion (an approximately 106-fold increase in the fraction of plasmid-bearing cells over 2 weeks, compared with a maximum of only 10-fold over 3 days in their study). Second, we specifically demonstrated the positive effects of nutrients and spatial structure on plasmid invasion. Together, our studies show the importance of the highly efficient conjugation systems of IncP-1 plasmids in the sweep of unselected traits, such as drug resistance, through bacterial populations.
A novel and intriguing observation in our study was that plasmid invasion was promoted by regular disturbance and random reorganization of the spatial population structure. Although the plasmid had invaded 13% of the population after 15 days in the ‘undisturbed filter transfer’ experiment (Figure 1a), 98% of all cells were invaded by the plasmid when the spatial structure was regularly disturbed (‘disturbed filter transfer’ protocol; Figure 1b). There are at least two possible explanations for this difference. The first involves the amount of contact between plasmid-bearing cells and plasmid-free cells in the population. In the undisturbed filter transfer protocol, the cells were in fixed positions. Therefore the plasmid could only transfer radially from the original donor cells to surrounding recipients, which in turn became transconjugants that could further transfer the plasmid to adjacent recipient cells. Thus, isolated clusters of recipients not in contact with the donor or transconjugant cells might remain plasmid-free. As the bacteria were spread over the filter with a cell spreader, cell distribution was uneven and gaps between cell clusters were observed (Figure 5). In the experiments with daily disturbance of the spatial structure, the cells were redistributed across the filter every day, and hence donors and transconjugants were more likely to be surrounded again by plasmid-free recipients, which constituted the majority during the first several days. Through this process, more plasmid-bearing cells had access to plasmid-free cells than in a situation with no regular disturbance. Microscopic analysis of our invasion experiments (Figure 5) showed that (1) as expected, the pattern of plasmid spread from the few original foci was a radial process and (2) the regular disturbance of spatial structure indeed increased the rate and extent of plasmid invasion.
A second possible explanation for the difference in invasiveness between undisturbed and disturbed transfer protocols is the difference in the location of cells relative to the agar and air interfaces. Random redistribution of cells on a daily basis results in different cells being closer to the agar surface or the air–colony interface, than in the previous 24 h period. Over time, this may give all cells the same probability to reside in a location that is most conducive to plasmid transfer. There are, of course, other possible factors influencing the results. For example, in the undisturbed filter transfer protocol, it is possible that accumulation of toxins or signalling molecules led to suppression of plasmid transfer, whereas these compounds would be removed daily in the disturbed filter transfer protocol. Moreover, in the latter protocol, the physical structure of the colony was altered daily by disrupting the extracellular polymeric matrix and possibly changing the intercellular distances, and cells were subjected to physical stresses due to resuspension by vortexing, centrifugation and replating on new filters. The effect of such stress factors on conjugation efficiencies has not been investigated, but recent studies have shown the importance of stress response mechanisms in conjugative gene transfer (Beaber et al., 2003; Zahrl et al., 2007). One additional caveat might be a slight loss of cells during every transfer due to the cell spreading procedure, which may affect plasmid dynamics. Although these alternative explanations should not be ignored, we believe that the first and second interpretations of the observed differences in plasmid invasiveness between undisturbed and disturbed population architecture are more plausible and are, in fact, supported by our model simulations (see below).
To further explore the factors mediating plasmid invasion in spatial populations, we expanded our earlier spatially explicit 2D lattice model (Krone et al., 2007) by incorporating enough 3D structure to simulate features of the experiments that appear to be essential in determining the extent of plasmid invasion. The mathematical results were consistent with the experimental results and support our intuition that plasmid invasion depends strongly on a combination of both access to nutrients and the geometry of contact between plasmid-containing and plasmid-free cells. The simulations nicely captured, in a way that was not over-sensitive to changes in parameters, the difference in invasiveness between the disturbed and undisturbed protocols. The simulations of the disturbed filter transfer protocol showed a final fraction of transconjugants that was slightly higher than in the corresponding experiments (Figures 1e versus b). We were able to get an even better fit by adjusting parameters that control conjugation rate and nutrient access, but this led to an underestimation of transconjugants in the undisturbed transfer protocol. In other words, we were not able to use one set of parameters to fit all the experimental results simultaneously with the accuracy we would have liked—although the qualitative patterns are unmistakable. This could be due to the simplifying assumptions in the model. However, further exploration of the model suggests that there may be an interesting biological explanation. In the simulations, disturbed transfer resulted in complete mixing (that is, randomization of cell locations). This, in principle, would maximize the potential for conjugation. In the experiments, on the other hand, this mixing step was implemented for 1 min by using a vortex mixer. Previous studies (Achtman, 1975) suggest that up to 5 min of vigorous agitation may be necessary to completely disrupt mating aggregates in E. coli. Although it would have been too far beyond the scope of our current study to include modelling of mating aggregates, we were able to simulate some of the effects of decreased mixing by running simulations that disturbed the spatial structure of the population only every other transfer. This, in fact, resulted in an almost perfect fit to the experimental data when using the same parameters as in Figure 1 (simulation data not shown). This is an example of how mathematical modelling can lead to testable hypotheses and thus help drive empirical work. Future work will include testing the effects of different vortexing times (representing various degrees of aggregate disruption) on plasmid invasion.
In general, one of the purposes of mathematical modelling is to help transform empirical data into understanding. By probing the sensitivity of the model to changes in parameters, we were able to note the generality of the results and explore the specific mechanisms most responsible for generating the observed behaviour. For example, the robustness of the qualitative behaviour in the simulations to changes in certain parameters (Figure 3) strongly suggests that the empirical observations are not specific to IncP plasmids and E. coli hosts, but rather should be applicable to any plasmid–host system that share certain basic features. By determining which combinations of parameter values did not lead to a fit of simulations to experiments (Figure 3), we were able to isolate the importance of different mechanisms. In additional simulations (data not shown), we found that both spatial limitations to nutrient diffusion (parameter m) and time of nutrient availability (parameter L) were essential in capturing the observed behaviour. For example, if we assumed all cells were active during the period of nutrient availability (m=40), then a given set of parameters either overestimated the ‘disturbed’ case or underestimated the ‘undisturbed’ case, but could not fit both cases simultaneously.
A few remarks regarding the model assumptions are as follows. (1) The main reason for the assumption of a local carrying capacity (M) was to match the approximate levelling-off of population size in the experiments. (2) One could argue that during the first day of growth, there are fewer cells and hence nutrients should be used up more slowly than on succeeding days. Although we could have chosen different values of L for these two scenarios, we had no direct measurements that would have rendered such choices less ad hoc. In addition, as the population reaches the carrying capacity, most nutrient consumption goes toward cell maintenance, not growth. Thus, it could well be that a constant value of L is a reasonable assumption. (3) The assumption of a constant conjugation rate for cells in the active state is in accord with some previous empirical work on surface-grown cultures, showing that little energy is required for plasmid transfer and, above a threshold activity level, transfer is not limited by metabolic activity (Normander et al., 1998; Hausner and Wuertz, 1999) (see also Figure 3). This is an area that is far from being resolved, however, and results may be influenced by strains, experimental conditions and even the methods used to infer transfer rates (Smets et al., 1993). (4) As discussed earlier, even the large 2000 × 2000 lattices correspond to a significantly smaller area than the filters used in the experiments. Thus, scalability of the model is important. We checked this by comparing simulations on 200 × 200 and 2000 × 2000 lattices and found that the results were consistent.
In conclusion, this study shows that under certain environmental conditions, plasmids can invade a plasmid-free population in the absence of selection for plasmid-encoded traits, but spatial structure and nutrients are essential to this invasive ability. This is consistent with observations that spatially structured, nutrient-rich habitats, such as the gastrointestinal tract, manure-treated soils, wastewater treatment plants, rhizo- and phylosphere and other surfaces that occasionally receive high supplies of nutrients, are hotspots for horizontal gene transfer (van Elsas and Bailey, 2002). For planktonic bacteria, the chance of contacting another bacterial cell is rare, unless cells densities are high (>1010 c.f.u. ml−1). But as bacteria adhere to surfaces, form close associations with other bacteria and have available nutrients, conjugation events can be much more favoured. A better understanding of the factors that promote plasmid invasion may eventually allow us to control the horizontal spread of desirable traits such as pollutant degradation, or undesirable phenotypes such as multidrug resistance, in bacterial communities. Our approach toward exploring these factors was to combine laboratory experiments with mathematical models and simulations. The combination of experimental and theoretical results provided insights that are more far ranging than what can be inferred from either approach alone.
References
Achtman M . (1975). Mating aggregates in Escherichia coli conjugation. J Bacteriol 123: 505–515.
Adamczyk M, Jagura-Burdzy G . (2003). Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim Pol 50: 425–453.
Bahl MI, Hansen LH, Licht TR, Sorensen SJ . (2007a). Conjugative transfer facilitates stable maintenance of IncP-1 plasmid pKJK5 in Escherichia coli cells colonizing the gastrointestinal tract of the germfree rat. Appl Environ Microbiol 73: 341–343.
Bahl MI, Hansen LH, Sorensen SJ . (2007b). Impact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol Lett 266: 250–256.
Beaber JW, Hochhut B, Waldor MK . (2003). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427: 72–74.
Bergstrom CT, Lipsitch M, Levin BR . (2000). Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155: 1505–1519.
Bouma JE, Lenski RE . (1988). Evolution of a bacteria/plasmid association. Nature 335: 351–352.
Bradley DE, Taylor DE, Cohen DR . (1980). Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol 143: 1466–1470.
Christensen BB, Sternberg C, Andersen JB, Eberl L, Moller S, Givskov M et al. (1998). Establishment of new genetic traits in a microbial biofilm community. Appl Environ Microbiol 64: 2247–2255.
Christensen B, Sternberg C, Molin S . (1996). Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (GFP) from Aequorea victoria as a marker. Gene 173: 59–65.
Costerton JW, Lewandowski Z, De Beer D, Caldwell D, Korber D, James G . (1994). Minireview: biofilms, the customized microniche. J Bacteriol 176: 2137–2142.
De Gelder L, Ponciano JM, Joyce P, Top EM . (2007). Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153: 452–463.
De Gelder L, Vandecasteele FP, Brown CJ, Forney LJ, Top EM . (2005). Plasmid donor affects host range of promiscuous IncP-1β plasmid pB10 in an activated-sludge microbial community. Appl Environ Microbiol 71: 5309–5317.
Dieckmann U, Law R, Metz JAJ . (2000). The Geometry of Ecological Interactions: Simplifying Spatial Complexity. Cambridge University Press: Cambridge, UK.
Dröge M, Pühler A, Selbitschka W . (1999). Horizontal gene transfer among bacteria in terrestrial and aquatic habitats as assessed by microcosm and field studies. Biol Fertil Soils 29: 221–245.
Dröge M, Pühler A, Selbitschka W . (2000). Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol Gen Genet 263: 471–482.
Ghigo JM . (2001). Natural conjugative plasmids induce bacterial biofilm development. Nature 412: 442–445.
Häagensen JAJ, Hansen SK, Johansen T, Molin S . (2002). In situ detection of horizontal transfer of mobile genetic elements. FEMS Microbiol Ecol 42: 261–268.
Hausner M, Wuertz S . (1999). High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol 65: 3710–3713.
Heuer H, Fox R, Top EM . (2007). Frequent conjugative transfer accelerates adaptation of an IncP-1 plasmid to an unfavourable Pseudomonas putida host. FEMS Microb Ecol 59: 738–748.
Krone SM, Lu R, Fox RE, Suzuki H, Top EM . (2007). Modeling the spatial dynamics of plasmid transfer and persistence. Microbiology 153: 2803–2816.
Lejeune P, Mergeay M, Van Gijsegem F, Faelen M, Gerits J, Toussaint A . (1983). Chromosome transfer and R-prime plasmid formation mediated by plasmid pULB113 (RP4::mini-Mu) in Alcaligenes eutrophus CH34 and Pseudomonas fluorescens 6.2. J Bacteriol 155: 1015–1026.
Levin BR, Lenski ER . (1983). Coevolution in bacteria and their viruses and plasmids. In: Fuyuma DJ, Sltakin M (eds). Coevolution. Sinauer Associates: Sunderland, Massachusetts, pp 99–127.
Levin BR, Stewart FM, Rice VA . (1979). The kinetics of conjugative plasmid transmission: fit of a simple mass action model. Plasmid 2: 247–260.
Licht TR, Christensen BB, Krogfelt KA, Molin S . (1999). Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145: 2615–2622.
Lundquist PD, Levin BR . (1986). Transitory derepression and the maintenance of conjugative plasmids. Genetics 113: 483–497.
Mazodier P, Davies J . (1991). Gene transfer between distantly related bacteria. Annu Rev Genet 25: 147–171.
MacArthur RH, Wilson EO . (1967). The Theory of Island Biogeography. Princeton University Press: Princeton, NJ, USA.
Molin S, Tolker-Nielsen T . (2003). Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol 14: 255–261.
Normander B, Christensen BB, Molin S, Kroer N . (1998). Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris). Appl Environ Microbiol 64: 1902–1909.
Pinedo AC, Smets BF . (2005). Conjugal TOL transfer from Pseudomonas putida to Pseudomonas aeruginosa: effects of restriction proficiency, toxicant exposure, cell density ratios, and conjugation detection method on observed transfer efficiencies. Appl Env Microbiol 71: 51–57.
Proctor GN . (1994). Mathematics of microbial plasmid instability and subsequent differential growth of plasmid-free and plasmid-containing cells, relevant to the analysis of experimental colony number data. Plasmid 32: 101–130.
Sambrook J, Russell DW . (2001). Molecular Cloning: a Laboratory Manual. Cold Spring Harbour Laboratory Press: Cold Spring Harbour, New York, USA.
Schlüter A, Heuer H, Szczepanowski R, Forney LJ, Thomas CM, Pühler A et al. (2003). The 64 508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a wastewater treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology 149: 3139–3153.
Schlüter A, Szczepanowski R, Pühler A, Top EM . (2007). Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev 31: 449–477.
Sia EA, Roberts RC, Easter C, Helinski DH, Figurski DH . (1995). Different relative importances of the par operons and the effect of conjugal transfer on the maintenance of intact promiscuous plasmid RK2. J Bacteriol 177: 2789–2797.
Simonsen L . (1990). Dynamics of plasmid transfer on surfaces. J Gen Microbiol 136: 1001–1007.
Smalla K, Haines AS, Jones K, Krögerrecklenfort E, Heuer H, Schloter M et al. (2006). Increased abundance of IncP-1β plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1β plasmids with a complex mer transposon as the sole accessory element. Appl Environ Microbiol 72: 7253–7259.
Smets BF, Barkay T . (2005). Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat Rev Microbiol 3: 675–678.
Smets BF, Rittmann BE, Stahl DA . (1993). The specific growth rate of Pseudomonas putida PAW1 influences the conjugal transfer rate of the TOL plasmid. Appl Environ Microbiol 59: 3430–3437.
Sørensen S, Bailey M, Hansen L, Krower N, Wuertz S . (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol 3: 700–710.
Sota M, Tsuda M, Yano H, Suzuki H, Forney LJ, Top EM . (2007). Region-specific insertion of transposons in combination with selection for high plasmid transferability and stability accounts for the structural similarity of IncP-1 plasmids. J Bacteriol 189: 3901–3908.
Stanier RY, Palleroni NJ, Doudoroff M . (1966). The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43: 159–271.
Stewart FM, Levin BR . (1977). The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87: 209–228.
Thomas CM, Nielsen KM . (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3: 711–721.
Thomas CM, Smith CA . (1987). Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol 41: 77–101.
Top EM, Moënne-Loccoz Y, Pembroke T, Thomas CM . (2000). Phenotypic traits conferred by plasmids. In: Thomas CM (ed). The Horizontal Gene Pool. Hardwood Academic Press: Amsterdam, pp 249–285.
Turner PE . (2004). Phenotypic plasticity in bacterial plasmids. Genetics 167: 9–20.
van Elsas JD, Bailey MJ . (2002). The ecology of transfer of mobile genetic elements. FEMS Microbiol Ecol 42: 187–197.
Zahrl D, Wagner A, Tscherner M, Koraimann G . (2007). GroEL plays a central role in stress-induced negative regulation of bacterial conjugation by promoting proteolytic degradation of the activator protein TraJ. J Bacteriol 189: 5885–5894.
Acknowledgements
We thank S Sax, S Bassler and L Rogers for technical assistance; M Sota for construction of K12Nal::gfp; H Suzuki for assistance with the statistical analysis and the entire Top Lab for suggestions on this paper. This work was funded by Grant no. R01GM073821 from the National Institute of General Medical Sciences (NIGMS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the National Institutes of Health (NIH). Randal E Fox was also in part supported by an NSF GK-12 fellowship through NSF Grant DGE-0538660.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fox, R., Zhong, X., Krone, S. et al. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J 2, 1024–1039 (2008). https://doi.org/10.1038/ismej.2008.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2008.53
Keywords
This article is cited by
-
Droplet size and surface hydrophobicity enhance bacterial plasmid transfer rates in microscopic surface wetness
ISME Communications (2022)
-
Properties affecting transfer and expression of degradative plasmids for the purpose of bioremediation
Biodegradation (2021)
-
Linking the resistome and plasmidome to the microbiome
The ISME Journal (2019)
-
Stopping Untreatable Pathogen Infections Using Peptide Ligands to Sabotage Pathogenic Cell Surface Proteins
Molecular Biotechnology (2019)
-
Persistence and reversal of plasmid-mediated antibiotic resistance
Nature Communications (2017)