Abstract
Purpose:
Ehlers–Danlos syndrome (EDS) comprises a group of overlapping hereditary disorders of connective tissue with significant morbidity and mortality, including major vascular complications. We sought to identify the diagnostic utility of a next-generation sequencing (NGS) panel in a mixed EDS cohort.
Methods:
We developed and applied PCR-based NGS assays for targeted, unbiased sequencing of 12 collagen and aortopathy genes to a cohort of 177 unrelated EDS patients. Variants were scored blind to previous genetic testing and then compared with results of previous Sanger sequencing.
Results:
Twenty-eight pathogenic variants in COL5A1/2, COL3A1, FBN1, and COL1A1 and four likely pathogenic variants in COL1A1, TGFBR1/2, and SMAD3 were identified by the NGS assays. These included all previously detected single-nucleotide and other short pathogenic variants in these genes, and seven newly detected pathogenic or likely pathogenic variants leading to clinically significant diagnostic revisions. Twenty-two variants of uncertain significance were identified, seven of which were in aortopathy genes and required clinical follow-up.
Conclusion:
Unbiased NGS-based sequencing made new molecular diagnoses outside the expected EDS genotype–phenotype relationship and identified previously undetected clinically actionable variants in aortopathy susceptibility genes. These data may be of value in guiding future clinical pathways for genetic diagnosis in EDS.
Genet Med 18 11, 1119–1127.
Similar content being viewed by others
Introduction
Ehlers–Danlos syndrome (EDS) is a group of overlapping hereditary disorders of connective tissue (HDCT).1,2,3 Predominant clinical phenotypes include skin fragility, easy bruising, and joint hypermobility.2 As a result of generalized connective-tissue fragility, blood vessels and internal organs may also be affected to a variable extent. Vascular EDS patients can present as early as the first 2 weeks of life with aneurysm or rupture of large blood vessels, or sudden spontaneous rupture of the bowel or other intra-abdominal organs.2,3,4
EDS is classified clinically by the 1997 Villefranche nosology.1 The three major groups are classical, vascular, and hypermobility-type EDS; others, including kyphoscoliosis, dermatosparaxis, and arthrochalasia, are extremely rare.1,2 The central pathological abnormality affects collagen packing or stability, mechanisms traditionally based on certain microscopic, biochemical, and genetic abnormalities in collagen types I, III, and V. Most classical EDS cases are caused by mutations in one of two genes encoding collagen type V (COL5A1, COL5A2); vascular EDS is caused largely by mutations in COL3A1, encoding collagen type III, and the genetics of hypermobility-type EDS remains largely unresolved and is predicted to be heterogeneous.2,4 A number of other genes encoding extracellular matrix proteins (PLOD1, CHST14, FKBP14; RIN2; PRDM5, ZNF469, B4GALT7, SLC39A13) have been reported as causes of additional very rare EDS presentations.2,4 Because of phenotypic heterogeneity and clinical overlap between EDS types, clinical evaluation alone is often not definitive, and even after genetic testing the majority of EDS cases remain without a molecular diagnosis.1,2
Next-generation sequencing (NGS) technologies offer the potential for genetic testing in a variety of disorders, including EDS.5 Using a newly developed NGS-based panel, we screened a mixed cohort of EDS patients for variants in key collagen genes and certain other genes known to cause EDS or related connective-tissue disorders. We sought to ascertain whether this approach would increase the proportion of genetic diagnoses and improve understanding of the relationship between genotype and phenotype in EDS by comparing the results obtained by NGS with previous genetic testing of individual genes by Sanger sequencing.
Materials and Methods
Selection of EDS cases
One hundred seventy-seven unrelated patients with suspected EDS referred by specialist (tertiary) clinicians to the National EDS Diagnostic Service (London, UK) were recruited to the study. The patients were predominantly female (67%) and Caucasian (89%). The mean age of the cohort was 33.6 years (range 2–78). Because the service studied complex EDS phenotypes, and because of their prognostic importance, patients with vascular complications were preferentially recruited to the study. Clinical diagnoses upon first interview, based only on clinical features, were as follows: classical EDS (or overlapping syndrome), 12%; vascular EDS (or overlapping syndrome), 12%; EDS hypermobility type or benign joint hypermobility syndrome (BJHS), 43%; and kyphoscoliotic EDS, 2%. Seven percent of cases had a phenotype overlapping multiple EDS types not falling into one specific EDS type and are referred to hereafter as complex EDS. Twenty-four percent of cases had features predominantly outside the EDS spectrum, here called “other HDCT.” This term was used for patients with nonspecific features of HDCT, whereby the criteria for a specific EDS type were not adequately fulfilled and the patient did not have an arterial complication (for example, osteogenesis imperfecta and myopathies). Another group not falling into a specific EDS type but with a history of arterial complication (defined as one or more of aortic or peripheral arterial aneurysm, dissection or rupture, cerebral aneurysm/subarachnoid hemorrhage) was designated as “other HDCT (vascular).”
Clinical categorization was based on 1997 Villefranche criteria1 by specialist EDS clinicians (F.M.P., A.V., N.G., with combined experience of 51 years). Seventy-six relatives were phenotyped and recruited to the study for segregation analysis. All study subjects were recruited according to Ethics Protocol Reference 11/LO/0883 (West London Research Ethics Committee).
Characterization of clinical, biochemical, histological, and ultrastructural phenotype
Phenotypic data were derived from clinical notes, including diagnostic scores (according to the Beighton system, Villefranche criteria, Ghent nosology, and Sillence criteria),1,6 light and electron microscopy, and collagen protein analysis performed at the National EDS Diagnostic Service, London. Genetic testing results (by conventional Sanger sequencing) for the UK National Health Service were recorded for comparison.
Skin biopsy specimens taken from patients’ upper inner arm were used for collagen protein analysis and microscopy. Fibroblasts were cultured from skin biopsy specimens, with collagen labeling and sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis performed as described.7 During light microscopy, dermal tissue blocks stained with hematoxylin and eosin/elastin Van Gieson were assessed to determine changes in dermal thickness and collagen:elastin ratio. During electron microscopy, skin biopsy specimens were placed directly into 4% glutaraldehyde in 0.1 mol/l phosphate buffer and processed for routine electron microscopy as described previously. 8 Thin sections (stained with uranyl acetate and lead citrate) were examined for ultrastructural abnormalities of collagen fibril size and arrangement and appearance of fibroblast endoplasmic reticulum.
DNA extraction and targeted exon sequencing
Saliva samples were collected using the Oragene DNA kit (Genotek, Ontario, Canada). DNA from whole-blood samples was extracted using the QIAamp DNA Blood Midi kit (Qiagen, Venlo, The Netherlands). Two NGS assays were designed to sequence the exons and exon–intron boundaries of genes commonly associated with EDS and overlapping phenotypes: (i) collagen NGS panel, with 375 primer pairs for COL1A1, COL1A2, COL3A1, COL5A1, and COL5A2, and (ii) aortopathy NGS panel, with 363 primer pairs for FBN1, TGFBR1, TGFBR2, MYH11, ACTA2, SMAD3, and MYLK (see Supplementary Tables S1 and S2 online; Supplementary Materials online). Both NGS panels were used for every patient.
Multiplex PCR was performed using the Access Array (Fluidigm, South San Francisco, CA) in batches of 47 test samples and 1 negative control (water blank) as described.9 Ninety-six purified samples were pooled and sequenced on the MiSeq sequencer (Illumina, San Diego, CA) generating 150-bp paired-end reads.
Read mapping, variant calling, and annotation
The quality of sequencing reads was assessed with FastQC.10 Reads were mapped to the GRCh37/hg19 human reference sequence using BWA-MEM v0.7.2.11 Sequence reads were realigned around insertions/deletions and base call quality scores were recalibrated with GATK v2.6-5.12 Single-nucleotide variants and short insertions/deletions (up to 20 nucleotides) were called with GATK UnifiedGenotyper. Variants were hard-filtered to achieve minimum coverage at a variant of ≥100 reads and allele balance of >0.25. Variant annotation was performed with ANNOVAR version 2013Aug23.13 Reference transcripts of genes are shown in Supplementary Table S3 online.
Pathogenicity assignment
Synonymous variants, intronic variants located outside exon/intron boundaries, and SNVs with minor allele frequency >0.1% in the 1000 Genomes Project (phase II release) or the NHLBI exome sequencing project data sets (as available in the 2013Aug23 version of Annovar) were excluded from further analysis. Splice site–disrupting variants and nonsense and frameshift coding variants with allele frequency <0.1% were considered pathogenic. Substitutions of glycine residues at GlyXY repeats within collagen triple helical domains were considered pathogenic, as were other variants previously reported as pathogenic, in established variant databases (LOVD14 and HGMD15) unless biochemical (collagen protein analysis), phenotype, and/or segregation data suggested otherwise. Missense variants were classified according to American College of Medical Genetics and Genomics (ACMG) guidelines,16 with the special additional case of glycine substitutions in collagen helical domains, which have the effect of disrupting helix formation and are therefore generally considered to be pathogenic because of loss of function conferred by these variants. All other variants were classified as variants of uncertain significance (VUS) and were further categorized based on available evidence according to ACMG criteria. 16 Filtered variants were validated by Sanger sequencing and submitted to the LOVD database.14 A single false-positive variant was found in exon 1 of SMAD3 (ID 382) and was removed from further analysis.
Statistical analysis
χ2 and Fisher’s exact tests were used to test for significant differences in categorical variables between individuals in different genotype or phenotype groups. The Mann–Whitney test was used to assess statistical differences between in silico prediction scores.
Results
Clinical phenotype
Table 1 summarizes the clinical phenotype spectrum tabulated by initial clinical diagnosis. There was wide phenotypic heterogeneity within each diagnostic group, with patients frequently possessing overlapping Villefranche criteria suggestive of more than one diagnostic category. Eighteen patients distributed across diagnostic groups had marfanoid features (of which two had Ghent Marfan systemic scores ≥7). Four patients diagnosed as “other HDCT” had signs consistent with osteogenesis imperfecta or an overlapping phenotype.
Results of previous genetic testing by Sanger sequencing
The traditional route to genetic testing by Sanger sequencing is phenotype-driven, usually by sequencing one or two of the most likely causative genes, based on clinical and laboratory data.
The frequency of a detected pathogenic variant in the anticipated gene was 33% for cases initially diagnosed as classical EDS or an overlapping phenotype (7 pathogenic variants in COL5A1/2 of 21 patients tested), although the frequency increased to 58% if those with overlapping classical/hypermobility phenotypes were excluded. The frequency of pathogenic variants in COL3A1 in patients initially diagnosed with vascular EDS was 50% (11 of 22 patients tested). In the four patients with a clinical phenotype of EDS/OI, the frequency of COL1A1/2 pathogenic variants was 75% ( Table 2 ). Outside of the anticipated gene, further testing revealed a specific pathogenic DNA variant in only 7.8% (12/155). In addition to short variants (SNVs and short indels) in collagen genes that were considered pathogenic, a complex rearrangement was identified causing complete allelic loss of COL3A1 and COL5A2 (ID 444), and pathogenic variants were identified in TNXB (ID 67), FBN2 (ID 1125), and FKBP14 (homozygous, ID 822) (see Supplementary Table S4 online). Pathogenic or likely pathogenic variants were also identified in three other patients: in FBN1 (ID 66), TGFBR1 (ID 706), and SMAD3 (ID 382) (see Supplementary Table S4 online). Several incidental genetic abnormalities not thought to be contributory to EDS phenotype were also observed: a heterozygote VUS in TNXB (ID 79), an FLNA variant (ID 538), and a 0.1-Mb polymorphic deletion identified by karyotyping (ID 801) (see Supplementary Table S4 online).
Targeted NGS sequencing
Amplicons of the collagen panel achieved a mean coverage of 602× with 95.0% bases covered above 100×, whereas those of the aortopathy panel achieved a mean coverage of 926× per amplicon with 97.4% targeted bases covered by 100 reads or more.
NGS sequence data were analyzed blind to previous genetic testing. The mean numbers of variants called per sample were 12.7 (range 5–25) and 28.7 (range 9–51) with the collagen and aortopathy NGS panels, respectively. After variants were filtered by allele frequency and variant type, a total of 28 pathogenic variants, 4 likely pathogenic variants, and 22 VUS were identified ( Tables 3 and 4 ).
Pathogenic variants identified by NGS
In the 21 cases categorized initially as classical EDS or overlapping phenotypes, we identified pathogenic variants in nine cases (41%): seven in the COL5A1 or COL5A2 genes and two in COL3A1 ( Table 3 ). The first of these two (ID 417) had overlapping features of classical, hypermobility, and vascular EDS; the second (ID 636) had convincing clinical features entirely consistent with classical EDS (see Supplementary Table S5 online). In the 22 patients initially diagnosed with vascular EDS, 11 had pathogenic variants in COL3A1, and 1 with overlapping features of vascular and classical EDS had a pathogenic variant in COL5A1 (functional corroboration was not possible because the patient declined skin biopsy—ID 1088; see Supplementary Table S5 online). One of the patients with an initial diagnosis of hypermobility-type EDS (ID 824) carried a pathogenic COL5A1 variant ( Table 3 and see Supplementary Table S5 online). Of the patients with an initial diagnosis of other HDCT, three had pathogenic variants in COL1A1 (all three diagnosed clinically with EDS/OI overlap) and another three had pathogenic variants in FBN1 ( Table 3 ; see Supplementary Tables S5 and S6 online). Supplementary Table S7 online shows the phenotype details of cases with no identified variants.
Likely pathogenic variants and VUS identified by NGS
Of the 26 VUS identified, 22 were dispersed among either hypermobile or other HDCT patients ( Table 4 ). These included variants in COL1A1, TGFBR1, TGFBR2, and SMAD3, which we categorized as likely pathogenic ( Table 4 and see Supplementary Table S6 online). One of these patients (ID 893) carried a helical domain Arg>Cys variant in COL1A1, a number of which have been shown to be pathogenic in previous reports.23
Although two particular variants had previously been reported as pathogenic, we classified these as VUS in our patients ( Table 4 and see Supplementary Table S5 online): a COL1A2 variant17 (ID 629) due to nonsegregation in first-degree relatives and a COL3A1 C-propeptide variant18 (ID 655) because of lack of clinical or biochemical corroboration in the index case or her first-degree relative who also carried the variant. Each of two hypermobility EDS patients was found to have two separate VUS: ID 38 in COL3A1 and COL5A2 and ID 39 in COL1A2 and COL3A1 ( Table 4 ; and see Supplementary Table S4 online). One patient initially categorized clinically as having classical EDS (ID 636) carried an intronic VUS in COL5A2 as well as a pathogenic COL3A1 variant (see Supplementary Tables S4 and S5 online).
In silico predictions of missense variants detected by NGS
The majority of missense pathogenic variants detected by NGS had in silico predictive scores of, or close to, 1.0, whereas the scores for VUS were more variable and lower (mean score = 0.70, P < 0.0001 for SIFT; 0.67, P = 0.0005 for Polyphen2; 0.89, P = 0.006 for MutationTaster; 0.60, P < 0.0001 for MutationAssessor); a similar pattern was observed for predictions of evolutionary conservation (see Supplementary Table S8 online). The two variants that were previously reported as pathogenic but that we classified as VUS (IDs 629 and 655) had high in silico prediction scores (see Supplementary Table S8 online) but lacked pathological or segregation data to support pathogenicity. A helical Arg>Cys variant in COL1A2 (ID 1151) and three other VUS in COL1A1, COL3A1, and COL5A1 (IDs 39, 824, and 828) also had high in silico predictions (see Supplementary Tables S5 online). In the aortopathy panel, three variants—in SMAD3 (ID 382), TGFBR2 (ID 814), and TGFBR1 (ID 706)—also had high in silico predictive scores; these three variants were in functionally important domains and the clinical phenotypes of these patients strongly suggested pathogenicity of the variants (see Supplementary Tables S6 and S8 online).
Clinical, genetic, and pathological correlates of collagen gene variants
Most pathogenic DNA variants in collagen genes were associated with a corresponding collagen protein abnormality on SDS-PAGE, but protein abnormalities were much less common in VUS (81.8% vs. 11.1%; P = 0.006). A similar difference in light and electron microscopy abnormalities was observed when pathogenic variants were compared with VUS (76.9 vs. 21.1%; P = 0.02). There was no significant difference (P > 0.05) in the frequency of presenting vascular complications between pathogenic COL3A1 variants (38%) and those with variants of other genes or when compared with mutation-negative cases (31%) (see Supplementary Table S9 online). There was also no significant difference in Beighton score based on gene or variant type (P > 0.05).
Comparison with Sanger sequencing
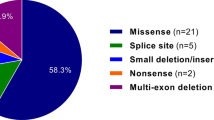
All of the 22 short pathogenic variants in the collagen genes and the pathogenic variants in FBN1 and SMAD3 that were identified by previous Sanger sequencing were also identified by the NGS panels ( Figure 1a ). Furthermore, the NGS panels newly identified seven pathogenic or likely pathogenic variants and 18 VUS that had not been detected by phenotype-guided Sanger sequencing ( Tables 3 and 4 ; Figure 1a ). The collagen NGS panel identified four unexpected pathogenic or likely pathogenic variants in genes that had not been selected for Sanger sequencing: (i) a COL3A1 variant (ID 636) in a patient previously diagnosed with classical EDS on the basis of clinical phenotype and the presence of an intronic, possible splice-disrupting COL5A2 variant (c.1402-10T>G); (ii) a loss-of-function COL5A1 variant in a patient with overlapping features of vascular and classical EDS phenotype (ID 1088) for whom only COL3A1 sequencing had been undertaken; (iii) an Arg>Cys variant in the helical domain of COL1A1 in a patient with a predominant hypermobility trait with fractures (ID 893); and (iv) a helical glycine disrupting COL5A1 variant in a patient with a predominant hypermobility trait (Beighton score 9/9) but minimal skin hyperextensibility (ID 824) (for details see Table 3 and Supplementary Table S5 online). In addition, six missense variants were identified with maximal or near-maximal in silico prediction scores for pathogenicity (see Supplementary Table S8 online). Because these variants did not meet ACMG criteria for pathogenicity or likely pathogenicity, they were classified as VUS. The aortopathy NGS panel identified four new variants not previously detected by clinical or genetic investigation, in FBN1, TGFBR1, and TGFBR2; two were pathogenic variants of FBN1 (IDs 766 and 378, both with initial diagnosis of other HDCT); one was a likely pathogenic variant (TGFBR2, ID 814, with an initial clinical phenotype of hypermobility-type EDS); and the fourth variant was classified as uncertain significance (TGFBR1, ID 475, with an initial clinical phenotype of other HDCT) (see Supplementary Table S6 online).
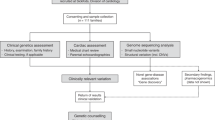
Alternative diagnostic pathways in EDS. (a) Yield of rare variants using traditional Sanger method (left) compared with the NGS panel (right). (b) Potential alternative clinical pathway to genetic diagnosis in EDS. Boxed numbers show the number of rare pathogenic variants and VUS identified in each group; percentages are the proportion of new pathogenic or likely pathogenic rare variants in our cohort of 177 EDS referrals. “Other” genetic abnormalities are those that were thought to underlie the EDS phenotype but are not covered by the NGS panel (three pathogenic variants in TNXB, FBN2, and FKBP14 and one large copy number variant; one VUS, a TNXB gene duplication). Dashed arrow indicates the potential for the NGS panel to become accessible to clinicians in secondary care. NGS, next-generation sequencing; EDS, Ehlers–Danlos syndrome; VUS, variants of uncertain significance.
Discussion
To our knowledge, this is the first report of the use of NGS to sequence a panel of relevant collagen and aortopathy genes in a large mixed cohort of patients with EDS or overlapping HDCT.
Our NGS panel was able to detect all previously identified short pathogenic variants in collagen and aortopathy genes and, in addition, seven newly identified pathogenic or likely pathogenic variants in COL1A1, COL3A1, COL5A1 (two cases), TGFBR2, and FBN1 (two cases), leading to new diagnoses in these patients. Of 18 newly detected VUS, a significant proportion had partial evidence of pathogenicity, based on previous reports, clinical phenotype, and in silico prediction score, including three in aortopathy genes (TGFBR1 and TGFBR2) and four in COL3A1, which, if shown to be pathogenic, alter clinical prognosis and management because of the associated high risk of arterial aneurysm, rupture, and/or bowel perforation.
Pathogenic collagen variants did not always correlate with the expected phenotype. In two patients we found pathogenic COL3A1 variants associated with a clinical phenotype of classical or overlapping classical EDS, one of whom (ID 636) also had an intronic COL5A2 variant, possibly splice site–disrupting, which could also contribute to his phenotype. Classical EDS is usually associated with pathogenic variants in COL5A1/2, but has rarely been associated with those in TNXB, COL1A1, or COL3A1.19,20 Pathogenic COL3A1 variants are normally considered diagnostic of vascular EDS, with reduced life expectancy from arterial and bowel complications.4,21
Because previous vascular EDS series have been selected mostly on the basis of clinical criteria (Villefranche) along with collagen type III protein deficiency or pathogenic COL3A1 variants, the frequency of other connective-tissue gene variants in clinical vascular EDS phenotypes is unknown.4,21 One of our patients clinically classified as having features predominantly of vascular EDS (ID 1088) showed a Leu1055X variant of COL5A1. One pedigree with a pathogenic COL5A1 variant segregating with a vascular EDS phenotype including arterial ruptures was recently reported (LOVD ID AN004203).14,22 Another patient (ID 824) for whom a pathogenic COL5A1 variant was found had an initial diagnosis of hypermobility-type EDS because of extensive hypermobility, soft but not hyperextensible skin, and family history of sudden cardiac death in a second-degree relative; functional corroboration was not possible for this patient because of loss to follow-up. As far as we are aware, such a phenotypic pattern has not been observed with a pathogenic COL5A1 variant, probably because COL5A1 sequencing to date has mostly been performed in phenotypically classical cases.2,3,19
Nine VUS with high in silico prediction scores or other evidence for pathogenicity were observed. These included one Arg>Cys variant within the helical domain of COL1A2 (ID 1151, p.Arg1338Cys) (see Supplementary Tables S4 and S5 online). Arg>Cys variants at helical locations in COL1A1 have been reported previously as predisposing to arterial fragility and other phenotypes (see Supplementary Table S5 online).23 Although the COL1A2 variant may be pathogenic, little evidence exists at the present time to establish its pathogenicity.
We also observed four variants in aortopathy genes TGFBR1, TGFBR2, and SMAD3 for which there was moderate or supporting evidence of pathogenicity (see Supplementary Table S6 online). Three of these were considered likely pathogenic: two in the functionally active kinase domains of TGFBR1 (ID 706) and TGFBR2 (ID 814) known to be associated with the majority of Loeys–Dietz syndrome variants to date24 and the third adjacent to the MH2 domain of SMAD3 (ID 382), which is known to regulate TGF-β signaling.25 Coronary artery dissection (seen in our patient) and other vascular complications have previously been caused by SMAD3 variants.26 The fourth variant, in TGFBR1 (ID 475), was in a patient with other HDCT, bone fragility, and a systolic murmur (and normal COL1A1/2). This variant was reported previously as a VUS associated with bicuspid aortic valve,27 and bone fragility is a recent addition to the LDS clinical spectrum28; therefore, this variant may underlie some of our patient’s features, but the evidence is less clear.
We reclassified two variants with high in silico prediction scores as VUS, although they were previously reported as pathogenic. The COL1A2 Arg708Glu variant detected in ID 629 was reported previously in a Marfanoid patient,17 but segregation analysis revealed hypermobility in all seven of the patients’ offspring, hypermobility in her (unrelated) husband, multiple fractures in one of the two offspring who carried the variant, and no fractures in the remaining five offspring, leading us to classify this variant as a VUS. The COL3A1 p.Lys1313Arg variant detected in ID 655 was previously reported in two unrelated patients with vascular EDS.18 However, collagen microscopy and biochemistry were normal in the patient, and neither the patient nor her sister, who also carried the variant, had features of vascular EDS, and structural studies indicated that the variant was unlikely to impair C-propeptide-mediated helical winding.29,30
Consistent with previous reports, we observed significant phenotypic overlap between our EDS diagnostic categories and other HDCT, as well as overlap among patients with pathogenic variants in individual causative genes.2,3,6,19,20,22,23 These factors point to the limitations of phenotype-driven genetic testing of individual candidate genes. The NGS approach presented here permits wider genetic testing than is possible with traditional Sanger sequencing, allowing more comprehensive genetic diagnosis. This led to new and revised diagnoses of patients in our cohort and widened the phenotypic spectrum ascribed to individual genes.
Based on our previous experience, and taking the results from this study as a whole, some general recommendations can be made. For cases with phenotypes overlapping classical or vascular EDS, the yield of pathogenic variants is high in the normally associated gene (COL3A1, COL5A1, and COL5A2), and occasionally one may identify pathogenic variants in another fibrillar collagen gene. Cases of uncomplicated hypermobility-type EDS with no family history of vascular complication had a low yield of pathogenic variants and a large number of VUS (most of which will not be fed back); NGS panel testing for such cases is unlikely to be diagnostic. Conversely, potential connective-tissue-disorder patients with a history of vascular complication, marfanoid features, or a significant family history should undergo NGS panel testing for collagen and aortopathy genes because our results indicate a reasonable likelihood of identifying pathogenic variants in one of these genes. Similarly, patients with EDS or other HDCT overlapping with osteogenesis imperfecta should also undergo collagen gene testing to exclude pathogenic COL1A1 or COL1A2 variants.
The interpretation of missense variants includes correlation with the complete clinical phenotype. In keeping with ACMG guidelines, we classify variants that are supported by some evidence of pathogenicity (e.g., high in silico scores and presence in functionally active domains) or associated with clinical features such as aortopathy as “likely pathogenic,” and we recommend that they be followed clinically with vascular imaging, familial segregation studies, and available structural or biochemical studies, including electron microscopy or protein analysis. Initial genetic counseling for such patients should point out that the true significance of the variant will not be known until these additional tests are complete. Although in the short-term some of these VUS may add uncertainty to the diagnostic process, we believe this is greatly outweighed by the prospect of making specific genetic diagnoses that would otherwise go undetected and form the basis of preventive screening in relatives. In the longer term, assignment of pathogenicity is likely to be facilitated by data from ongoing large-scale genome sequencing projects in patient and control cohorts.
We used the PCR-based Fluidigm Access Array followed by Illumina MiSeq. Mean coverage per amplicon was greater than 140× for all amplicons, which is comparable to results with previously reported assays using this methodology.9,31 Although this method is unable to detect large deletions or chromosomal rearrangements, these are rare in EDS.2 In addition, although false-positive variant calls have previously been reported with this approach,31 we detected only a single variant (in exon 1 of SMAD3) that could not be confirmed by Sanger sequencing. Because NGS methods have higher throughput, achieve greater coverage, and may be more cost-efficient than conventional Sanger sequencing,31 they may be applicable to wider phenotypic groups, for which this study may serve as a guide to future genetic testing. We suggest that the higher throughput, lower cost, and comparable or greater diagnostic yield of NGS could alter the optimal diagnostic pathway such that genetic testing with NGS panels can occur earlier in the diagnostic pathway, prior to tertiary or quaternary clinical appraisal ( Figure 1b ).
Disclosure
T.J.A. has received speaker honoraria from and has research collaborations with Illumina and has received consultancy fees from AstraZeneca. The other authors declare no conflict of interest.
References
Beighton P, De Paepe A, Steinmann B, Tsipouras P, Wenstrup RJ. Ehlers–danlos syndromes: revised nosology, Villefranche, 1997. Am J Med Genet 1998;77:31–7.
De Paepe A, Malfait F. The Ehlers–Danlos syndrome, a disorder with many faces. Clin Genet 2012;82:1–11.
Proske S, Hartschuh W, Enk A, Hausser I. [Ehlers–Danlos syndrome—20 years experience with diagnosis and classification at the university skin clinic of Heidelberg]. J Dtsch Dermatol Ges 2006;4:308–318.
Pepin M, Schwarze U, Superti-Furga A, Byers PH. Clinical and genetic features of Ehlers–Danlos syndrome type IV, the vascular type. N Engl J Med 2000;342:673–680.
Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet 2010;11:31–46.
Loeys BL, Dietz HC, Braverman AC, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet 2010;47:476–485.
Nuytinck L, Narcisi P, Nicholls A, Renard JP, Pope FM, De Paepe A. Detection and characterisation of an overmodified type III collagen by analysis of non-cutaneous connective tissues in a patient with Ehlers–Danlos syndrome IV. J Med Genet 1992;29:375–380.
Ong KT, Plauchu H, Peyrol S, et al. Ultrastructural scoring of skin biopsies for diagnosis of vascular Ehlers–Danlos syndrome. Virchows Arch 2012;460:637–649.
Vandrovcova J, Thomas ER, Atanur SS, et al. The use of next-generation sequencing in clinical diagnosis of familial hypercholesterolemia. Genet Med 2013;15:948–957.
Babraham Bioinformatics. FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Cornell University Library. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. http://arxiv.org/abs/1303.3997.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38. doi:10.1093/nar/gkq603.
LOVD—Ehlers–Danlos Syndrome Variant Database. https://eds.gene.le.ac.uk/home.php.
The Human Gene Mutation Database (HGMD). http://www.hgmd.org/.
Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424.
Phillips CL, Shrago-Howe AW, Pinnell SR, Wenstrup RJ. A substitution at a non-glycine position in the triple-helical domain of pro alpha 2(I) collagen chains present in an individual with a variant of the Marfan syndrome. J Clin Invest 1990;86:1723–1728.
Pickup MJ, Pollanen MS. Traumatic subarachnoid hemorrhage and the COL3A1 gene: emergence of a potential causal link. Forensic Sci Med Pathol 2011;7:192–197.
Malfait F, De Paepe A. Molecular genetics in classic Ehlers–Danlos syndrome. Am J Med Genet C Semin Med Genet 2005;139C:17–23.
De Paepe A, Nicholls A, Narcisi P, et al. Ehlers–Danlos syndrome type I: a clinical and ultrastructural study of a family with reduced amounts of collagen type III. Br J Dermatol 1987;117:89–97.
Pepin MG, Schwarze U, Rice KM, Liu M, Leistritz D, Byers PH. Survival is affected by mutation type and molecular mechanism in vascular Ehlers–Danlos syndrome (EDS type IV). Genet Med 2014;16:881–888.
Monroe GR, Harakalova M, van der Crabben SN, et al. Familial Ehlers–Danlos syndrome with lethal arterial events caused by a mutation in COL5A1. Am J Med Genet A 2015;167:1196–1203.
Malfait F, Symoens S, De Backer J, et al. Three arginine to cysteine substitutions in the pro-alpha (I)-collagen chain cause Ehlers–Danlos syndrome with a propensity to arterial rupture in early adulthood. Hum Mutat 2007;28:387–395.
Loeys BL, Schwarze U, Holm T, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med 2006;355:788–798.
Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003;425:577–584.
van der Linde D, van de Laar IM, Bertoli-Avella AM, et al. Aggressive cardiovascular phenotype of aneurysms—osteoarthritis syndrome caused by pathogenic SMAD3 variants. J Am Coll Cardiol 2012;60:397–403.
Bonachea EM, Zender G, White P et al. Use of a targeted, combinatorial next-generation sequencing approach for the study of bicuspid aortic valve. BMC Med Genom 2014;7:56.
Kirmani S, Tebben PJ, Lteif AN, et al. Germline TGF-beta receptor mutations and skeletal fragility: a report on two patients with Loeys–Dietz syndrome. Am J Med Genet A 2010;152A:1016–1019.
Bourhis JM, Mariano N, Zhao Y, et al. Structural basis of fibrillar collagen trimerization and related genetic disorders. Nat Struct Mol Biol 2012;19:1031–1036.
Stembridge NS, Vandersteen AM, Ghali N, et al. Clinical, structural, biochemical and X-ray crystallographic correlates of pathogenicity for variants in the C-propeptide region of the COL3A1 gene. Am J Med Genet A 2015;167A:1763–1772.
Halbritter J, Diaz K, Chaki M, et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet 2012;49:756–767.
Morissette R, Schoenhoff F, Xu Z, et al. Transforming growth factor-beta and inflammation in vascular (type IV) Ehlers–Danlos syndrome. Circ Cardiovasc Genet 2014;7:80–88.
Vandersteen AM, Lund AM, Ferguson DJ, et al. Four patients with Sillence type I osteogenesis imperfecta and mild bone fragility, complicated by left ventricular cardiac valvular disease and cardiac tissue fragility caused by type I collagen mutations. Am J Med Genet Part A 2014;164A:386–391.
Acknowledgements
T.J.A. was supported by intramural funds from the MRC Clinical Sciences Centre, the National Institute for Health Research (NIHR) via the Imperial Biomedical Research Centre, the Imperial College British Heart Foundation Centre of Research Excellence, and the University of Edinburgh. R.W. was supported by an Imperial-Wellcome Trust Clinical Training Fellowship.
The authors thank all patients and families, the EDS National Diagnostic Service, Northwick Park Hospital, and London North West Healthcare NHS Trust. They are also grateful to Julie Leary, Carole Cummings, Marion Bartlett, Hilary Bell-Joseph, Claire Neuwirth, and Vera Weerasinghe for help with patient recruitment and clinical records and to Philip Sawle for collagen biochemical analysis.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Tables
(ZIP 220 kb)
Rights and permissions
About this article
Cite this article
Weerakkody, R., Vandrovcova, J., Kanonidou, C. et al. Targeted next-generation sequencing makes new molecular diagnoses and expands genotype–phenotype relationship in Ehlers–Danlos syndrome. Genet Med 18, 1119–1127 (2016). https://doi.org/10.1038/gim.2016.14
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.14
Keywords
This article is cited by
-
Gastric perforation leading to the diagnosis of classic Ehlers–Danlos syndrome: a case report
Journal of Medical Case Reports (2021)
-
A novel missense mutation of COL5A2 in a patient with Ehlers–Danlos syndrome
Human Genome Variation (2016)