Abstract
Purpose:
The purpose of this study was to evaluate the clinical utility of targeted exome sequencing (TES) as a molecular diagnostic tool for patients with skeletal dysplasia.
Methods:
A total of 185 patients either diagnosed with or suspected to have skeletal dysplasia were recruited over a period of 3 years. TES was performed for 255 genes associated with the pathogenesis of skeletal dysplasia, and candidate variants were selected using a bioinformatics analysis. All candidate variants were confirmed by Sanger sequencing, correlation with the phenotype, and a cosegregation study in the family.
Results:
TES detected “confirmed” or “highly likely” pathogenic sequence variants in 74% (71 of 96) of cases in the assured clinical diagnosis category and 20.3% (13 of 64 cases) of cases in the uncertain clinical diagnosis category. TES successfully detected pathogenic variants in all 25 cases of previously known genotypes. The data also suggested a copy-number variation that led to a molecular diagnosis.
Conclusion:
This study demonstrates the feasibility of TES for the molecular diagnosis of skeletal dysplasia. However, further confirmation is needed for a final molecular diagnosis, including Sanger sequencing of candidate variants with suspected, poorly captured exons.
Genet Med 18 6, 563–569.
Similar content being viewed by others
Introduction
Skeletal dysplasias are a heterogeneous group of genetic disorders of the skeletal system caused by mutations of genes involved in bone and cartilage metabolism.1 This condition is characterized by short stature, limb and spine maldevelopment or deformities, and precocious osteoarthritis. A recent update of the nosology and classification of genetic skeletal diseases included 40 groups and 456 disease entities, listing 226 causative genes that have been discovered for 316 diseases.2 The clinical diagnosis of the specific disease entity causing skeletal dysplasia can be difficult for several reasons. First, individual clinicians have limited experience with these rare diseases. Second, mutations in different genes can sometimes cause similar phenotypes, and mutations of the same gene can sometimes cause multiple phenotypes. Third, characteristic skeletal manifestations either tend to disappear after skeletal maturity or do not appear at a young age. Finally, some disease entities are not yet well established. Molecular diagnosis offers the possibility of identifying the specific entity underlying skeletal dysplasia, but it is challenging to determine which gene(s) should be tested despite a difficult clinical diagnosis.
Next-generation sequencing has led to rapid advances in human genomics and is being widely applied for clinical sequencing.3 In recent years, researchers have used whole-exome sequencing (WES) and whole-genome sequencing (WGS) techniques to identify novel genes causing skeletal dysplasia.4,5 However, WES and WGS are very expensive, and the number of variants detected by these techniques is too high for their application in routine genetic testing. In this context, screening of a certain number of genes by panel-based targeted exome sequencing (TES) could be a useful alternative because of its superior accuracy through high depth, simplicity of analysis, and relatively low costs compared with WES, WGS, and Sanger sequencing.6 Moreover, skeletal dysplasia represents an ideal disease condition for detection by TES; many diseases are caused by various mutations scattered along the vast number of genes of matrix proteins without hotspots, often making it unclear which gene should be tested.
We performed a literature review and selected 255 genes associated with skeletal dysplasia. Capture-based target gene enrichment and massively parallel sequencing were adopted to identify sequence variants. We recruited 185 patients over a 3-year period and studied genetic variations in these patients, including single-nucleotide variations, small insertions/deletions (indels), and copy-number variations (CNVs). We also investigated agreement between clinical diagnosis before TES and molecular diagnosis after TES to identify the clinical utility of TES as a diagnostic tool.
Materials and Methods
Subjects and clinical diagnosis
This study was approved by our institutional review board. We recruited 185 patients who were diagnosed with or suspected to have skeletal dysplasia based on clinical findings and radiographic survey ( Table 1 ). Informed consent was obtained from all patients; thereafter, genomic DNA was extracted from the circulating leukocytes from the proband and, when possible, his or her parents. The patients were divided into five categories according to the certainty of clinical diagnosis and the status of the prospective genotype ( Table 2 ). Patients with assured clinical diagnosis of a skeletal dysplasia for which one or only a few genotypes are known to be responsible, such as achondroplasia, were excluded from this study because they are not considered suitable candidates for TES. Patients with an assured clinical diagnosis with a single known causative gene were assigned to category I, and those with an assured clinical diagnosis with several known causative genes were assigned to category II. Category III included patients considered to have skeletal dysplasia of a certain category but for which a specific diagnosis of disease entity could not be made, for example, unspecified-type spondylometaphyseal dysplasia. Category IV included patients with dysplasia that has a possible genetic background. This category included 20 cases of Legg-Calvé-Perthes disease of (i) bilateral involvement, especially synchronous; (ii) familial occurrence; and (iii) suspicious but indefinite skeletal abnormalities such as epiphyseal hypoplasia of the contralateral hip or other joints and vertebral abnormalities. Finally, 25 patients with skeletal dysplasia with their genotypes confirmed by previous Sanger sequencing constituted category V, which was used to verify the sensitivity of TES.
Targeted exome sequencing
We designed RNA baits covering the 255 target genes using a custom capture array (SureSelect Customized Kit; Agilent Technologies, Santa Clara, CA). The gene list contained 168 genes featured in the 2010 revision of the nosology and classification system,2 in addition to 87 genes newly reported after publication of that revision. The baits included all exons of target genes, as determined by previous studies of skeletal dysplasia–related genes (Supplementary Table S1 online). We prepared a sequencing library from patient genomic DNA using the Agilent SureSelect Target Enrichment System protocol. TES was performed with 101-bp paired-end reads on an Illumina MiSeq platform (Illumina, San Diego, CA).
Identification and prioritization of pathogenic variants
Reads were aligned to the human genome reference sequence (hg19) using Burrows-Wheeler Aligner version 0.7.5 (ref. 7) with the MEM algorithm. We used SAMTOOLS version 0.1.18 (ref. 8), GATK version 2.4-7 (ref. 9), and Picard version 1.93 (http://broadinstitute.github.io/picard/) for sorting, indexing SAM/BAM files, local realignment, and duplicate markings. Base recalibration was performed using GATK (known single-nucleotide polymorphisms and indels from dbSNP137, Mills, and 1000 Genome Project gold-standard indels from b37 sites, and 1000 Genome Project phase 1 indels from b37 sites). To identify mutations from the targeted genes, sequence variants were called by Unified Genotyper in GATK and recalibrated by GATK based on dbSNP137, Mills indels, HapMap, and Omni. ANNOVAR10 was used to annotate the variants. We first selected exonic and splicing variants, including nonsynonymous variants and small indels. Variants with an allele frequency of more than 1% were discarded based on the National Heart, Lung, and Blood Institute’s ESP 6500, the 1000 Genomes Project, and our in-house database containing the exomes of 192 Korean individuals (manuscript and Web-based database in preparation). Finally, we correlated these variants with clinical and radiographic findings to determine the candidate pathogenic variant(s), which were additionally checked by a segregation test performed for each family. All the details regarding the workflow used in the study are shown in schematic form in Supplementary Figure S1 online. All variants labeled as candidates were confirmed by Sanger sequencing.
Molecular diagnosis using determined variants
Molecular diagnosis was performed by correlating the clinical and radiographic findings with candidate sequence variants obtained through TES. The status or certainty of molecular diagnosis was stratified into five categories. “Confirmed” was assigned if the sequence variants in the gene expected from the phenotype were either previously reported for the phenotype or indicated as pathogenic in functional assay(s). “Highly likely” was assigned if the candidate sequence variant(s) was not previously reported as associated with the phenotype but was predicted to be damaging by in silico analyses and occurred at a frequency of less than 1% in the normal population; this was further supported by a segregation test or well-known pathogenicity such as Gly substitution in the α-helix of collagen, a truncation mutation, or a previously reported mutation in the same amino acid. “Less likely” was assigned if the candidate sequence variant(s) in the gene expected from the phenotype was predicted to be benign by in silico analysis and was rather common among the normal population or showed contradictory segregation in the family. “Deferred” was assigned when further analysis was ongoing or parental DNA was not yet available. Finally, “no candidate” status was assigned when no sequence variants relevant to the phenotype were detected by TES.
Detection of CNV
We investigated CNVs using TES data for patients assigned the “no candidate” status and for whom CNV was considered a possible pathogenic mutation. We calculated the sequencing coverage for each exon region using the Depth of Coverage module in GATK. The depth of each exon was normalized, and the copy-number ratio was calculated by the normalized depth of each exon in 96 patients. To verify the results obtained from CNV analysis by TES, we performed array comparative genomic hybridization (aCGH) using the Unrestricted SurePrint G3 CGH Microarray 4x180K Kit (G4826A-022060; Agilent Technologies). The raw aCGH data were normalized using quantile and LOESS methods from the LIMMA package.11 To correct for X-chromosome mismatch between male and female patients, the median value for each probe was calculated and subtracted from the raw value of each probe. DNAcopy and SNPchip software packages12 were used for visualization.
Results
High performance of the TES method
We designed baits for the 255 genes known to cause genetic skeletal disorders (Supplementary Table S1 online). Mean coverage of the targeted exons was achieved at 105.31× (SD: 26.03), with 96.53% more coverage than that achieved at 10× (Supplementary Table S2 online). To explore the efficiency of target coverage, the coverage at a base-pair resolution was calculated for all intended target exons (3,910 exons) in the 255 target genes. About 4.3% (n = 168) of the targeted exons was calculated to have 90% coverage, and 27 exons were calculated to have zero coverage (Supplementary Table S3 online). The genomic regions with coverage less than 90% are presented in Supplementary Table S4 online. To characterize these exons further, we plotted them with GC content, which could have affected the mean coverage. The results revealed that most of the exons attained a mean coverage at 1, but the coverage for those with more than 60% GC content dropped dramatically. Zero-coverage exons were widely distributed in GC content (Supplementary Figure S2a online). In addition, an examination of the relationship between mean coverage and mean depth for all exons showed that a shallow mean depth corresponded to low mean coverage (Supplementary Figure S2b online). These results suggest that low m--ean coverage was the result of the shallow read depth caused by high GC content; however, high GC content does not directly cause zero-coverage exons.
Systematic prioritization of pathogenic variants
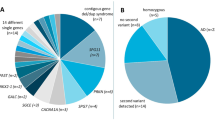
To prioritize potential pathogenic variants, we systematically applied several filtering steps (Supplementary Figure S1 online). Of the average 1,611 variants per patient, approximately 20 remained candidates at the final step. Candidate variants were correlated with clinical and radiological findings to select the candidate sequence variant that was most likely to be responsible for the phenotype and to make a molecular diagnosis. The prioritization process provided 128 candidate pathogenic sequence variants. Sanger sequencing revealed one (0.76%) as a false positive. Subsequent analysis of segregation and the database of previously reported mutations assigned 122 sequence variants in 109 cases (58.9%) as “confirmed” or “highly likely” pathogenic mutations for the phenotype, and 6 variants in 6 cases (3.2%) as “less likely” pathogenic mutations for the phenotype. In 70 cases (37.8%), “no candidate” sequence variant was identified. The 122 “confirmed” or “highly likely” sequence variants in 109 cases are summarized in Tables 3 and 4 and Supplementary Table S5 online. At the nucleotide level, the variants were 89 point mutations at exons, 10 point mutations at intron boundaries, 18 deletions of 1 to 13 nucleotides, and 5 insertions of 1 to 4 nucleotides. At the protein level, they were predicted to produce 82 amino acid substitutions, 36 truncated proteins, and 4 deletions.
Assured clinical diagnosis (categories I and II)
The 96 patients had an assured clinical diagnosis of varying numbers of possible pathogenic mutations (categories I and II) ( Table 2 ). In those categories, 71 patients (74.0%) were found to harbor “confirmed” and/or “highly likely” pathogenic sequence variants ( Table 3 , Supplementary Table S5 online), and 2 (2.1%) were found to harbor “less likely” pathogenic sequence variants. In the remaining 23 patients (24.0%), “no candidate” pathogenic sequence variant was identified. Among the 23 “no candidate” cases, one turned out to have “highly likely” pathogenic sequence variants in a recently identified pathogenic gene that was not included in the TES gene panel. A woman with type III osteogenesis imperfecta was found to harbor compound heterozygous mutations of WNT1 (ref. 13) via WES conducted after TES. For another patient, a novel candidate causative gene was identified after TES and is now under further investigation.
Uncertain clinical diagnosis (categories III and IV)
Categories III and IV included 64 cases of uncertain clinical diagnosis. Thirty-five were considered to have skeletal dysplasia, but a specific diagnosis could not be made because their phenotypes were ambiguous or they presented after skeletal maturity or when they were too young. The remaining 29 cases were suspected to have skeletal dysplasia without certainty. In this uncertain clinical diagnosis category, 13 patients (20.3%) were found to harbor “confirmed” and/or “highly likely” pathogenic sequence variant(s) ( Table 4 ), and 4 (6.3%) were found to harbor “less likely” variants. Forty-seven patients (73.4%) were assigned as “no candidate.” Their clinical diagnoses included Legg-Calvé-Perthes disease (14 cases), spondyloepiphyseal dysplasia–unspecified (7 cases), spondylometaphyseal dysplasia–unspecified (6 cases), sclerosing bone disease (3 cases), and mesomelic dysplasia (3 cases).
Known genotype (category V)
In contrast to the patients in categories I–IV, we included 25 cases with genotypes already known through previous Sanger sequencing to verify the sensitivity of TES. The cases harbored 30 mutations in 15 genes. The mutations consisted of 22 missense point mutations at exons, 1 point mutation at an intron boundary, 5 deletions of 1 to 13 nucleotides, and 2 insertions of 4 and 21 nucleotides. Details of the variants are presented in Supplementary Table S5 online.
Complementary genomic screening for candidate variants
For the patients categorized as “no candidate” by TES, we performed CNV analysis. Based on the read-depth approach,14 one patient who was clinically diagnosed with mesomelic dysplasia had the lowest read depth of the SHOX gene among all other patients with skeletal dysplasia, indicating that she had a SHOX deletion (Supplementary Figure S3a online). This finding was confirmed by aCGH of the patient’s X chromosome (Supplementary Figure S3b online). Polymerase chain reaction experiments also confirmed the lack of SHOX in this patient compared with two other patients (Supplementary Figure S3c online). The clinical diagnosis was Leri-Weill syndrome caused by SHOX mutation;15 hence, the result of the CNV analysis using TES was concordant with the clinical diagnosis.
Discussion
Molecular and genetic diagnosis using next-generation sequencing technology has become widely adopted in medicine. A panel-based TES approach to identify pathogenic mutations therefore serves as an efficient diagnostic tool.16 The US Food and Drug Administration recently approved several next-generation sequencing–based in vitro diagnostic tests for clinical purposes.17 Here, the merits of TES for molecular diagnostics were tested in 185 patients with skeletal dysplasia: 160 patients with unknown genotypes and 25 patients with known genotypes. TES detected “confirmed” or “high likely” pathogenic sequence variants in 84 of 160 untested patients (52.5%) and successfully detected all of the pathogenic sequence variants (100%) that were previously reported.
The utility of TES could depend on the patients’ clinical diagnosis status. If the clinical manifestation is known to be one with only a single or few known pathogenic mutation(s), Sanger sequencing for the specific mutation, not TES, would be the optimal test to confirm the molecular diagnosis. Achondroplasia and infantile cortical hyperostosis (Caffey disease) are examples of such manifestations. However, in cases that have multiple candidate variants in a single or multiple candidate causative gene(s) (categories I and II), even though a clinical diagnosis is assured, TES has advantages over Sanger sequencing because it can test for all candidate genes simultaneously. Osteogenesis imperfecta, multiple epiphyseal dysplasia, spondyloepiphyseal dysplasia congenita, and other type II collagenopathies belong to these categories. This study revealed that TES detected “confirmed” or “highly likely” pathogenic mutations in 71 of 96 patients (74.0%) in this category.
Another strength of TES is its ability to provide a molecular diagnosis when the clinical manifestation does not suggest any specific disease (category III), or even when it is not certain whether the patient has a genetic skeletal disorder (category IV). Here, TES provided a molecular diagnosis for 13 patients whose specific clinical diagnoses could not be made by expert diagnosticians ( Table 4 ). Because the clinical and radiographic characteristics of skeletal dysplasia usually become less conspicuous after skeletal maturity, sometimes at a very young age, molecular confirmation for mutations in diverse genes is a powerful tool for diagnosis. Prenatal diagnosis of skeletal dysplasia is a prominent example of when a specific clinical diagnosis is difficult to make not only because of a lack of ossification in the skeletal system but also difficulty in conducting comprehensive radiographic studies at this stage.18 Furthermore, certain forms of skeletal dysplasia show diverse phenotypes, making clinical diagnosis difficult or controversial, even by specialists.19 Interestingly, 7of 13 cases in which a pathogenic mutation in categories III and IV was identified by TES were later confirmed to be type II collagenopathy. These were clinically diagnosed cases of multiple epiphyseal dysplasia (category II), spondyloepiphyseal dysplasia–unspecified (category III), or Legg-Calvé-Perthes disease (category IV) ( Table 3 ). This reflects the phenotypical diversity of type II collagenopathy, and also demonstrates the robustness of TES.
Surprisingly, the success rate of detecting pathogenic sequence variation was only 17.1% in category III, which was even lower than that in category IV. Data from patients with skeletal dysplasia, for which the causative gene has not been discovered, had been collected for a period of time to conduct WES. These patients were included in this study as category III to exclude the known genes, which we believe might contribute to the low success rate in this category. They were of an spondylometaphyseal dysplasia–unspecified type, mesomelic dysplasia–unspecified type, and so on. Screening for these causative genes can not only enable a fast and efficient molecular diagnosis but also identify cases that do not have any mutations in known genes. Such cases would constitute a good study cohort for targeting a novel causative gene. Seven of 29 cases (24.1%) that were not definitively skeletal dysplasia were found to have pathogenic sequence variations among the gene panel. Four cases, thought to be Legg-Calvé-Perthes disease, were found to be of type II collagenopathy, and the remaining three patients were very young or had a very mild phenotype; therefore, there was an equivocal suspicion of skeletal dysplasia before TES.
Sule et al.20 reported a next-generation sequencing platform allowing simultaneous sequencing of genes that cause inherited disorders of high or low bone mineral density. They used a platform similar to ours, with 34 genes, and reported benefits and limitations of such technologies that we also experienced. To apply TES in clinical use, cost-effectiveness needs to be considered when designing the size of the gene panel, based on the scope of disorders to be covered, the number of patients to be tested in a given time, and the unit cost. In our clinical and laboratory settings, a panel of 255 genes, including all skeletal dysplasia genes, was optimal.
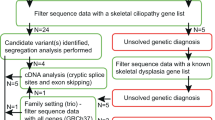
We could not identify any candidate variant in the 70 patients using our TES method, for several possible reasons. First, some might not have had a genetic disorder. For example, 20 patients with Legg-Calvé-Perthes disease were included in this study because some reports have indicated that a type II collagenopathy manifests as this entity.21,22 However, most of them did not show any sequence variant through TES. Second, some patients might have had skeletal dysplasia due to a causative gene that has not yet been discovered and is therefore not on the TES gene list. Those cases are candidates for WES or WGS. Because we designed sequencing baits to capture only genes previously reported to be associated with skeletal dysplasia, pathogenic variants in an unknown gene possibly escaped detection. For example, one patient with osteogenesis imperfecta in whom no pathogenic mutation was identified by our TES gene panel turned out to have “highly likely” pathogenic sequence variations in WNT1, which was reported as one of the causative genes for this phenotype after diagnosis by TES in this study.13 Third, some genetic defects are simply not detectable by TES using an exon-only capture strategy. They include deep intron variants, variants in regulatory/enhancer sequences, indels of nucleotides larger than a certain size, and chromosomal anomalies.23,24 For such patients, additional analysis might involve WES, WGS, aCGH, multiplex ligation-dependent probe amplification, or fluorescence in situ hybridization ( Figure 1 ).
Schematic workflow used in this study. Recruited patients were classified into five categories based on the certainty of clinical diagnosis and the status of the prospective genotype. The patients in category I were screened by a single gene test, and those in categories II to V were screened by skeletal dysplasia–targeted exome sequencing (TES). Sequence variants obtained by TES were prioritized using a bioinformatics analysis along with correlation with clinical and laboratory findings, and a segregation test was performed for the variants. We assume that “confirmed” and “highly likely” variants were pathogenic in nature unless alternative methods were required to identify novel variants.
Of the 70 cases in which “no candidate” sequence variant was identified, one showed a shallow read-depth ratio for all the exons in SHOX, which was compatible with the clinical phenotype. We subsequently confirmed a deletion of SHOX using aCGH. This case showed the detection of CNVs using TES. However, it has often been reported that most tools identifying CNVs based on the results of TES, particularly by the depth-of-coverage approach, do not show high performance for the detection of CNVs,25 and additional experiments should confirm them. Not only uneven capture efficiency but also noncontiguous TES target regions make it difficult to accurately determine CNVs and can generate false-positive results.
Those features originate from the technical limitation of TES. For example, GC content is known to affect the efficacy of target coverage.26 During preparation of the library, polymerase chain reaction is used to amplify target regions; however, high GC content reduces the efficiency of such amplification.27 Moreover, the hybridization of capture probes to the target sequences can be hindered by high or low GC content28 (Supplementary Table S4 online). In our study, we confirmed that both sequencing depth and coverage were dependent on GC content (Supplementary Figure S2 online). We recommend that the coverage depth for those exons be confirmed, followed by Sanger sequencing, if they are clinically suspected to harbor any pathogenic variant. In addition to GC content, homologous regions in the genome can influence capture efficiency. In the presence of pseudogenes, capture efficiency might be lower or the pseudogene per se might be captured. This outcome would trigger false-positive variant calls based on misalignment of the sequences.3 Repetitive sequences of several major gene families (e.g., the collagen genes) also often result in such mapping issues. High-depth sequencing or selecting reads with good mapping quality might address those issues, and Sanger sequencing for all detected variants in such exons will be necessary.
Conclusion
We developed and applied TES for diagnosing 185 patients with or suspected to have skeletal dysplasia in order to screen 255 genes known to be involved in the pathogenesis of skeletal dysplasia. The clinical utility of TES was demonstrated in the detection of pathogenic sequence variants of single-nucleotide polymorphisms, small indels, and possibly CNVs in the patients with skeletal dysplasia. TES is particularly useful when multiple genes or multiple sites of a huge gene need to be simultaneously sequenced to reach a correct molecular diagnosis. Our results support the use of TES as a diagnostic platform. Nonetheless, the limitations of this technique and the range of mutations that it can cover should be taken into consideration in interpreting the results. Selected candidate variants and exons that are suspected but poorly captured on TES screening should be tested using Sanger sequencing.
Disclosure
The authors declare no conflict of interest.
References
Spranger JW, Brill PW, Nishimura G, Superti-Furga A, Unger S. Bone Dysplasia: An Atlas of Genetic Disorders of Skeletal Development, 3rd edn. Oxford University Press: New York, 2012.
Warman ML, Cormier-Daire V, Hall C, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A 2011;155A:943–968.
Rehm HL, Bale SJ, Bayrak-Toydemir P, et al.; Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Commitee. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013;15:733–747.
Min BJ, Kim N, Chung T, et al. Whole-exome sequencing identifies mutations of KIF22 in spondyloepimetaphyseal dysplasia with joint laxity, leptodactylic type. Am J Hum Genet 2011;89:760–766.
Faqeih E, Shaheen R, Alkuraya FS. WNT1 mutation with recessive osteogenesis imperfecta and profound neurological phenotype. J Med Genet 2013;50:491–492.
Park JH, Kim NK, Kim AR, et al. Exploration of molecular genetic etiology for Korean cochlear implantees with severe to profound hearing loss and its implication. Orphanet J Rare Dis 2014;9:167.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760.
Li H, Handsaker B, Wysoker A, et al.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164.
Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 2006;7:252.
Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007;23:657–663.
Fahiminiya S, Majewski J, Mort J, Moffatt P, Glorieux FH, Rauch F. Mutations in WNT1 are a cause of osteogenesis imperfecta. J Med Genet 2013;50:345–348.
Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics 2013;14(suppl 11):S1.
Munns C, Glass I. SHOX-related haploinsufficiency disorders. In: Pagon RA, Adam MP, Ardinger HH, et al (eds.). GeneReviews. University of Washington: Seattle, WA, 1993.
Consugar MB, Navarro-Gomez D, Place EM, et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med 2015;17:253–261.
Bijwaard K, Dickey JS, Kelm K, Tezak Z. The first FDA marketing authorizations of next-generation sequencing technology and tests: challenges, solutions and impact for future assays. Expert Rev Mol Diagnost 2014:1–8.
Parilla BV, Leeth EA, Kambich MP, Chilis P, MacGregor SN. Antenatal detection of skeletal dysplasias. J Ultrasound Med 2003;22:255–8; quiz 259.
Zankl A, Jackson GC, Crettol LM, et al. Preselection of cases through expert clinical and radiological review significantly increases mutation detection rate in multiple epiphyseal dysplasia. Eur J Hum Genet 2007;15:150–154.
Sule G, Campeau PM, Zhang VW, et al. Next-generation sequencing for disorders of low and high bone mineral density. Osteoporos Int 2013;24:2253–2259.
Miyamoto Y, Matsuda T, Kitoh H, et al. A recurrent mutation in type II collagen gene causes Legg-Calvé-Perthes disease in a Japanese family. Hum Genet 2007;121:625–629.
Li N, Yu J, Cao X, et al. A novel p. Gly630Ser mutation of COL2A1 in a Chinese family with presentations of Legg-Calvé-Perthes disease or avascular necrosis of the femoral head. PLoS One 2014;9:e100505.
Koboldt DC, Ding L, Mardis ER, Wilson RK. Challenges of sequencing human genomes. Brief Bioinform 2010;11:484–498.
Shigemizu D, Fujimoto A, Akiyama S, et al. A practical method to detect SNVs and indels from whole genome and exome sequencing data. Sci Rep 2013;3:2161.
Kadalayil L, Rafiq S, Rose-Zerilli MJ, et al. Exome sequence read depth methods for identifying copy number changes. Brief Bioinform 2015;16:380–392.
Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 2009;27:182–189.
Aird D, Ross MG, Chen WS, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol 2011;12:R18.
Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res 2000;28:4552–4557.
Acknowledgements
The authors express their deep appreciation to Jürgen Spranger, Christine Hall, Gen Nishimura, Andrea Superti-Furga, and Sheila Unger for their continued discussion in making clinicoradiographic diagnosis of many cases included in this study. The study was supported by a grant from the Korean Health Technology Research and Development Project, Ministry of Health & Welfare, Korea (HI12C0014), and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning (NRF-2013R1A2A2A01067398).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Figure S1
(PPT 144 kb)
Supplementary Figure S2 and S3
(PPT 367 kb)
Supplementary Tables
(DOC 504 kb)
Rights and permissions
About this article
Cite this article
Bae, JS., Kim, N., Lee, C. et al. Comprehensive genetic exploration of skeletal dysplasia using targeted exome sequencing. Genet Med 18, 563–569 (2016). https://doi.org/10.1038/gim.2015.129
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2015.129
Keywords
This article is cited by
-
Diagnostic yield of rare skeletal dysplasia conditions in the radiogenomics era
BMC Medical Genomics (2021)
-
Estimation of the carrier frequencies and proportions of potential patients by detecting causative gene variants associated with autosomal recessive bone dysplasia using a whole-genome reference panel of Japanese individuals
Human Genome Variation (2021)
-
Novel loss-of-function variants of TRAPPC2 manifesting X-linked spondyloepiphyseal dysplasia tarda: report of two cases
BMC Medical Genetics (2019)
-
A novel mutation in the C-propeptide of COL2A1 causes atypical spondyloepiphyseal dysplasia congenita
Human Genome Variation (2017)