Abstract
Purpose:
The practice of “genomic” (or “personalized”) medicine requires the availability of appropriate diagnostic testing. Our study objective was to identify the reasons for health systems to bring next-generation sequencing into their clinical laboratories and to understand the process by which such decisions were made. Such information may be of value to other health systems seeking to provide next-generation sequencing testing to their patient populations.
Methods:
A standardized open-ended interview was conducted with the laboratory medical directors and/or department of pathology chairs of 13 different academic institutions in 10 different states.
Results:
Genomic testing for cancer dominated the institutional decision making, with three primary reasons: more effective delivery of cancer care, the perceived need for institutional leadership in the field of genomics, and the premise that genomics will eventually be cost-effective. Barriers to implementation included implementation cost; the time and effort needed to maintain this newer testing; challenges in interpreting genetic variants; establishing the bioinformatics infrastructure; and curating data from medical, ethical, and legal standpoints. Ultimate success depended on alignment with institutional strengths and priorities and working closely with institutional clinical programs.
Conclusion:
These early adopters uniformly viewed genomic analysis as an imperative for developing their expertise in the implementation and practice of genomic medicine.
Genet Med 16 12, 954–961.
Similar content being viewed by others
Introduction
Interest in the integration of “genomic medicine” into routine medical care is rapidly increasing, given the association of genetic variation with disease risk, development and progression of somatic diseases and of cancer, and drug efficacy or risk of adverse events.1 The core technology is termed “next-generation sequencing” (NGS), broadly defined as gene sequencing using massively parallel strategies. The first publication on the use of this technology was in 2005,2 in which sequencing of 25 million bases at 99% or better accuracy in one 4-hour run was able to achieve an approximately 100-fold increase in throughput over existing Sanger sequencing technology. Nine years later, NGS has found applications in sequencing the genome, exome, transcriptome, and microbiome, with more than 6,000 citations in PubMed using this technology and >100 publications accruing per month.3
Initial citations consisted of single reports of NGS being applied to diagnose conditions for which conventional molecular methods had failed4 or for which known diseases (usually cancer) were resistant to established pharmacologic therapies.5 Broader case-based experience is now forthcoming that supports the concept that genomic testing can benefit patients with undiagnosed disorders whose management has exhausted all standard care options.6 Nevertheless, expansion of NGS utilization for more routine use still requires establishment of an evidence base for most clinical applications.7,8 Achieving this goal will require access to extensive information about patient genotype, clinical phenotype, treatment outcomes, and the economics of delivering health care informed by genomic information.9 Regardless, having “genomic medicine” available now is increasingly becoming a mark of leading health systems,10 based on the premise that efficiency of health-care delivery and quality of life will be improved for their patients.11,12
The decision to bring NGS and genomic medicine into the clinical armamentarium is challenging because the actual clinical evidence base for systematic use of this technology is not firmly established. Such decision making is made more difficult by substantial uncertainty in how use and interpretation of this technology will be paid for, both by federal programs and by private payers.13 Obtaining regulatory approval for clinical application of this technology, as through Clinical Laboratory Improvement Amendments certification, is also a barrier that to date only a few laboratories have cleared.
The Personalized Health Care Committee of the College of American Pathologists is tasked with examining the role of new technologies for molecular diagnostics and advising the College on policies and programs to support the implementation of such technologies. A seven-person workgroup was commissioned to examine the business arguments for clinical use of NGS. Given the lack of published information regarding institutional implementation of NGS, the workgroup endeavored to identify the reasons for health systems choosing to bring NGS into their clinical laboratories and to understand the process by which such decisions were made. Interviews of the laboratory directors for institutional “early adopters” of NGS technologies were conducted. The interviews covered reasons for establishing NGS testing within a health system (as opposed to using a reference laboratory), the process for making such a decision, barriers to bringing such testing in-house, lessons learned, and key drivers of success. This study is complementary to a recently published study that examined challenges and barriers in instituting genomic medicine from the perspective of institutional clinical leadership.14 Taken together, these studies provide valuable information for current and future institutional leaders, laboratory and clinical alike, considering establishment of genomic testing programs.
Materials and Methods
Selection of participants
“Early adopter” health systems were identified by the following criteria: (i) having members serving on the College of American Pathologists “Personalized Health Care Committee”; (ii) performing NGS testing for human diagnostics under a Clinical Laboratory Improvement Amendments license; (iii) a record of peer-review publication documenting the use of massively parallel sequencing for human laboratory diagnostics; and/or (iv) institutional public announcements of NGS implementation. We excluded for-profit laboratories or independent laboratory networks based on the premise that there was an inherent business argument for their performing NGS testing as reference laboratories. Rather, health systems were considered to constitute the “end users” of genomic information and, in choosing to perform NGS testing themselves, faced the more complex decision-making process. By these inclusion criteria, interviews of pathologist leaders from 13 academic medical centers were conducted. All prospects accepted the invitation to be interviewed.
Survey instrument
A 1-hour telephone interview was designed, using open-ended questions to give the interviewees an opportunity for discussion. Approximately 1 week before the interview, a standardized list of 10 questions was sent to the interviewee. All interviews were conducted by one interviewer (J.H.K.) and were recorded. The 10 core questions were covered in the interview; for the purposes of elaboration, the core questions were supplemented by subquestions during the interview (the full set of questions and subquestions is given in Supplementary Table S1 online). This adaptive process allowed us to gain deeper insights into our core question areas.
Data compilation and analysis
The complete interview transcripts were redacted to remove information that could potentially identify a given institution or individual. The word count for each institutional response was used as an initial measure of the information content,15,16 with results presented in Table 1 . Data were then analyzed according to qualitative content analysis.17,18 This methodology is appropriate for qualitative studies using open-ended interviews because it enables identification and quantification of specific concepts mentioned by respondents.19,20,21
Results
Survey metrics
Thirteen institutions fulfilled one or more of the inclusion criteria; these were located in 10 different states and one Canadian province. Two of the US institutions had Clinical Laboratory Improvement Amendments certification for clinical use of NGS technologies. Table 1 shows the rank order of word count for the 10 interview questions. There was no statistical difference in word count (data not shown) in relation to whether the individual being interviewed was the medical director of the molecular diagnostics laboratory (six interviews) or a pathology department chair (five interviews) or if the department chair was invited to join the medical director in the interview (two interviews).
The greatest information content pertained to prospective department and institutional decision making (question 1); only limited information was given about strategies for measuring outcomes (question 9). This relative lack of content in response to the latter is telling because institutions had to make their decisions about implementing NGS in the absence of a priori evidence about whether doing so would help them achieve improved metrics for more efficient health care and/or better patient outcomes. That being said, respondents spoke about (i) the ability to identify druggable targets in cancer; (ii) elimination of redundant testing; (iii) earlier diagnosis; (iv) growth in laboratory test volumes and expansion of testing menus; (v) achieving “better educated” clinicians and pathologists; (vi) increased interactions between the clinical laboratory and treating clinicians; (vii) assessment of provider satisfaction with laboratory support of patient care; and (viii) achievement of a self-sustaining financial model.
Clinical applications and intended time line for deployment
Ten institutions indicated cancer genomics to be the primary clinical application for NGS testing. For the remaining three institutions, medical genetics was the primary application. Four institutions currently offered NGS testing clinically, seven institutions were looking to deploy NGS testing within the next 6 to 12 months, for one institution the time line for deployment was uncertain, and one institution used send-out testing to offer genomic testing. Regardless of the initial intent, five of the seven institutions that gave estimates of relative target case volumes assumed that they would be performing NGS testing for both cancer genomics and medical genetics. The remaining two assumed that 100% of their NGS testing would be for cancer genomics. One institution also planned on branching out into NGS testing for infectious disease diagnostics.
Drivers for implementation
Table 2 shows the reasons given for deciding to perform clinical genomic testing, which are listed in order of decreasing frequency of mention. Institutions frequently gave multiple reasons. Clinical demand and anticipated testing efficiency were almost uniform reasons. A critical indicator was genomic testing that was already being “sent out” to a reference laboratory, which could be redirected to an in-house facility if established. This argument was even more compelling as the number of different tests being ordered increased, constituting the reason for performing one larger gene panel in-house. Because the testing platform is not gene or disease specific, testing could be consolidated. For only one institution was recruitment of oncologists (“cancer folks”) mentioned as a reason for deploying NGS sequencing. None of the respondents mentioned retention of personnel as a reason for bringing NGS testing in-house.
The institutional decision-making process
In 10 of the 13 institutions, officers above the level of department chair were involved in review and approval of the decision to implement NGS testing, up to the level of the university president (one institution) and chief operating officer of the hospital (five institutions). In two institutions, the dean of the college of medicine was a key decision maker, and in one institution, the cancer center director. In only three institutions the chair of the department of pathology was the senior institutional officer making the decision.
Institutional funding
Twelve institutions commented on the sources of funding for bringing NGS in-house. For five institutions, 100% of the funding was from the hospital. For five other institutions, funding was entirely from the department of pathology, either entirely as research funds (one department) or as a mixture of research and department funds (four departments). It was for three of these latter five institutions that the decision-making process had been entirely controlled by the pathology department chair. In one institution, the department, hospital, and cancer center contributed equally to the implementation of NGS testing; this was the institution that involved the cancer center director in the decision-making process. Finally, one Canadian institution noted that funding for their NGS initiative was, ultimately, governmental because the Canadian government supports both research and the clinical delivery system.
Interactions with payers
For the four institutions already offering NGS testing, each had established mechanisms to work with payers for reimbursement; one institution also relied heavily on reimbursement through “institutional accounts” in which the client institutions paid the NGS testing laboratory through reference laboratory agreements. These four respondents noted that agreements between their institution and specific payers or client institutions were specific to their local environment and that these might not be applicable to other institutions.
Potential competitive value
Twelve of the 13 institutions were firm in their conviction that the availability of NGS testing constituted a competitive market advantage, both for their institutions as a whole and for their cancer centers. For four institutions, expansion of geographic reach of their health system constituted an additional perceived value. As one respondent stated, NGS testing was viewed as having a “halo effect” on the overall stature of the institution and its cancer center. The last institution considered that the clinical utility of NGS testing was not sufficiently established, so that the competitive value was as yet uncertain.
The question can be asked of whether competitive market advantage is an appropriate justification for implementing NGS testing. A starting point for this issue is performance of NGS testing in-house. “The whole reason for doing this is to be able to provide the same or often better service than the single gene test, at substantially lower costs.” Further, “In our practice environment, the standard of care for certain malignancies [can be met] faster and cheaper by NGS, than by doing those panels of genes by Sanger sequencing.” “It is not about the sequencing machine. It is about the cost-per-test.” Beyond that, and even drawing on only case-based experience, “avoidance of futile therapeutic regimes is a powerful argument for the use of NGS testing.” This translates into oncologists “no longer planning treatments based on one particular gene test, but on a number of different genetic abnormalities.”
Institutional expectations
Only four respondents commented on institutional expectations for outcomes as factoring into the decision-making process. These were enrolling patients in clinical trials (two institutions) and obtaining grant funding (one institution). The fourth institution did not expect financial sustainability for at least 5 years.
Barriers to implementation
Although only one survey question asked about “lessons learned” (question 8), sprinkled throughout the interview transcripts were comments about encountering and overcoming barriers to NGS implementation and encountering unanticipated needs ( Table 3 ). A recurrent theme in the interviews was the challenge of having adequate institutional expertise in informatics. The two institutions with large existing research programs in genomics obtained valuable mentorship in starting up the bioinformatics for clinical genomics. Two other institutions were successful in recruiting experts in both genomic testing and informatics. Such recruitment was acknowledged as a significant barrier to overcome because “these people are in demand, and they will have competing job offers.” These two institutions spoke in glowing terms about the positive effect of these experts on the institution. By contrast, two other institutions noted that they obtained institutional funding for equipment acquisition but not the resources for recruitment, staff training, or equipment validation in order to get NGS operational. Not surprisingly, these two institutions were not yet operational for their NGS testing.
Lessons learned
Each institution offered its own lessons learned, with substantive overlap among institutions ( Table 4 ). Common themes were that implementation took or was taking longer, and required more thorough involvement of clinical stakeholders, than anticipated. Four institutions started with a commercially established testing application before expanding into a broader test menu. Ironically, even for cancer-oriented institutions, germ-line testing for medical genetics was a plausible initial application for NGS so as to gain initial expertise and validate testing platforms. A related recommendation was to make sure that the chosen NGS testing aligned well with institutional strengths in clinical expertise, research programming, and, if available, informatics. This would help ensure that clinical case volumes would justify the performance of relevant NGS testing in-house. A second institution reported on the need to have a multidisciplinary advisory group to establish clear guidelines for which patients should be offered NGS testing as a lesson learned.
In the respondents’ words, these are additional thoughts about achieving success:
-
“If you are a small place, wait for (i) the platforms, informatics, and reporting to be worked out, and (ii) for research to demonstrate that [NGS] is clinically useful and superior. [From an implementation standpoint], you are better off if you can wait until NGS is pre-packaged, but then you will not be a leader in the field.”
-
“If you are going to be a leader, the greatest barrier is optimizing the design of the gene panel for the clinical application. These panels must be tested and validated before clinical implementation.”
-
“It is a lot easier to generate the data than [to] interpret it. You have to adequately resource the informatics function for interpretation. If you don’t, you end up with a very expensive paper weight in your department.”
-
“It is important to be adequately capitalized when you start. Trying to build NGS in a piecemeal fashion is not a successful strategy.”
-
“You must do high quality DNA extractions.”
-
“You have to work closely with clinical colleagues, so as to avoid offering something that will not be useful to their clinical practice. Match your platform with the testing that is needed.”
-
“Start small. Build your volume gradually so that you keep your quality up.”
-
“Understand the limitations of the technology. For testing specific small regions of the genome, NGS may not be the preferred technology.”
-
“Decide what data will be reported. Will you report information that is not being tested for?”
Measurement of outcomes
Intended evaluation of the outcomes of instituting NGS testing was qualitative in nature, along the lines of better management of patients (five institutions) and earlier clinical diagnosis (two institutions). When mentioned, intended quantitative measures included growth in test volumes, expansion in test menus, and elimination of other laboratory testing (four, two, and one institutions, respectively). Surveys were planned to assess physician and/or patient satisfaction (one institution each). In two institutions, achievement of a financially sustainable model was viewed as a key outcome measure.
Assistance from organized pathology
The respondents gave strong suggestions for contributions that could be made by the College of American Pathologists. Education of the professional and technical workforce for the performance of NGS testing was most frequently mentioned (seven institutions), followed by core activities of the College’s Laboratory Accreditation Program (testing standards, validation protocols, checklists, and proficiency testing). Dissemination of information about coding, billing, and reimbursement, as well as about national data networks and databases was mentioned by more than one institution.
Strategies for implementation
Three general strategies emerged for the implementation of NGS testing in-house: (i) overlaying NGS testing on established institutional polymerase chain reaction–based testing, before expanding the test menu; (ii) beginning with commercially validated NGS testing platforms, before expanding the test menu; (iii) starting with customized (but limited) NGS panels for specific applications, before expanding the test menu. An alternative to these is making the decision ab initio to outsource NGS testing. In this last instance, a derivative decision is then whether to have the genomic analysis also performed on an outsourced basis or to perform the informatics analysis in-house. The relevant value of each starting point is determined by local institutional needs and priorities. In turn, the timing and scope of expansion in available NGS test menus and applications will be determined in part by the success of the initial implementation and by the continued evolution of institutional strategies and priorities.
Proposed flowchart for institutional decision making and implementation
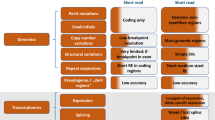
These survey responses permit the construction of a proposed flowchart for the institutional process of bringing NGS testing in-house ( Figure 1 ). Although each institution will have its own unique requirements, this flowchart attempts to capture the essential elements for institutional decision making and project management.
Discussion
The institutional decision to implement massively parallel sequencing, as exemplified by NGS, presents both an opportunity and a challenge. Because NGS testing is performed by a clinical laboratory, we surveyed the laboratory leaders, who carry ultimate responsibility for the validity of such testing. Whether medical director of a molecular diagnostic laboratory or department chair, these individuals are pathologists: physicians who are experts in the performance, interpretation, and clinical application of laboratory diagnostics. For these respondents, institutional demand and the challenges of implementation were the primary issues to be faced.
As a dominant initiating theme, demand for NGS testing from clinical colleagues most often originated with oncologists. An equally dominant theme was the expectation that massively parallel NGS testing would, ultimately, be more efficient than single test–based methodologies. That being said, a critical early decision was whether to implement custom panels or to implement commercially available testing products. The former permits tailoring of the panels to specific institutional needs; the latter enables implementation of testing that already has been validated elsewhere. Recommendations were made in favor of each starting strategy.
Returning to the question of whether “competitive advantage” or “recognition” is a justifiable reason for implementing NGS, there are a number of convergent considerations. Synthesizing their many responses, the institutions choosing to implement NGS sequencing (i) wished to attract patients to their institution; (ii) have both outstanding clinicians and laboratorians for the practice of genomic medicine; (iii) contribute to new knowledge about the efficacy of NGS testing in patient care; and (iv) control costs, testing algorithms, and the quality of NGS testing. In the eyes of these institutions, acknowledging that “recognition” and “competitive advantage” might also be forthcoming is not so much justification as important collateral for being an institution that advances innovation in health care.
The reported barriers and challenges in implementing NGS testing begin with having insufficient institutional expertise in informatics and in the technical performance of testing. Additional issues included the need for test validation, competent performance and interpretation of the testing, determination of test utility, and financing, both of the initial implementation and of the ongoing testing program. Collectively, these issues reflect the unique requirements imposed by the regulatory environment for performing clinical testing22 as opposed to testing that is done for research purposes.23 These barriers and challenges are distinctly different from the institutional challenges reported by Manolio et al.,14 which were limited evidence or consensus for which genomic variants were medically relevant; lack of reimbursement for genomically driven clinical interventions (which is different from concern about reimbursement for the performance of the genomic testing); and burden to patients and clinicians not only in assaying and reporting of testing but also in intervening and following up on genomic findings.
There was commonality in one challenge found by us and reported by Manolio et al.,14 which for our study respondents was the amount of data that needed to be curated and for respondents in the Manolio study was the need for a framework to define and catalog clinically actionable genomic variants. Key for establishing such a framework is a knowledge base that captures sequence variants and their phenotypic associations.14,24 It is therefore notable that a separate workgroup of the College of American Pathologists Personalized Healthcare Committee is currently examining the principles and specifications for “clinically actionable” genomic databases, and the actual creation and curation of such databases is a pan-industry effort. That being said, the presence of such databases is only an intermediate step. Professional interpretation of genomic variants found during testing on individual patients must occur, and it is this step that constitutes the practice of medicine. The finalization of a test report requires both access to appropriate genomic databases and judgment of the clinical importance of test findings for the individual patient.
Regarding reimbursement for NGS testing, the four laboratories already performing NGS testing had established reportedly productive conversations with payers, and six other laboratories in the process of implementation were optimistic about communication with payers. One institution had well-established payment mechanisms through interinstitutional agreements. With that one exception, institutions were not expecting financial sustainability of their NGS testing in the near future, and consumption of institutional resources, including research funds, was a necessary requirement for ongoing performance of such testing. This simple fact underscores the difficulty facing institutions in deciding whether or not to implement NGS testing.
This report highlights the challenges of instituting NGS testing, in the form of the need for additional institutional resources, and the diversity of unexpected issues. A theme that overlaps with the Manolio study is the highly multidisciplinary nature of implementing genomic testing.14 A corollary is identified in this study: the NGS testing chosen should align with institutional strengths both in clinical programming and in research initiatives. We note that, in more than 38,000 words of interview content, the word “governance” was mentioned only twice, once as an unanticipated additional requirement for implementation and once as a lesson learned after implementation. We have incorporated the requirement for governance as a requirement ab initio for implementing genomic medicine ( Figure 1 ). We also recommend that the patient voice be represented in such a governance mechanism because genomic medicine not only cuts across technical practice and medical management but also invokes ethical and societal issues.6,25
The limited information content for assessment of outcomes was striking. Establishment of quantitative metrics is immediately possible for test volume, test menu, and financial sustainability, and it is achievable for physician satisfaction and patient satisfaction through survey mechanisms. However, the actual assessment of health-care outcomes remains elusive. The relative paucity of institutional specifications for assessing outcomes challenges the establishment of an evidence base for the presumed clinical utility of genomic medicine. We consider setting up the metrics prospectively for quantitative assessment of patient and population outcomes essential for institutional deployments of NGS technologies. Reporting on such metrics, including whether implementation of NGS creates new problems in delivering effective patient care, should remain a priority for future studies.
This study was not designed to determine whether the decision to implement NGS testing was actually sound. Second, for these “early adopters,” metrics for outcomes were not yet available. Rather, in our interviewing this first wave of NGS adopters, their real-time experiences can instead serve as a guide for other institutions or health-care entities giving consideration to the value of incorporating genomic medicine into their clinical programming, in advance of an extensive evidence base being established for the ultimate utility of genomic medicine. By also creating an extensive lexicon of key concepts, this study helps advance a recommendation by Manolio et al.14 that institutional efforts to establish genomic medicine programs should benefit from collaboration using a more structured sharing of best practices. In doing so, this study helps delineate the key elements of decision making and implementation, and it can serve as a guide for future institutions and health systems that are considering offering genomic testing to their patient populations.
Disclosure
J.P. is a cofounder of PierianDx, founded in December 2013 after completion of this study. The other authors declare no conflict of interest.
References
Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 2008;452:872–876.
Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;437:376–380.
www.ncbi.nlm.nih.gov/pubmed/. “Next Generation Sequencing” or “NGS” as Search Terms. Accessed 29 March 2014.
Dimmock DP, Trapane P, Feigenbaum A, et al. The role of molecular testing and enzyme analysis in the management of hypomorphic citrullinemia. Am J Med Genet 2008;146A:2885–2890.
Jones SJM, Laskin J, Li YY, et al. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol 2010;11:R82.
Jacob HJ, Abrams K, Bick DP, et al. Genomics in clinical practice: lessons from the front lines. Sci Transl Med 2013;5:194cm5.
Crawford JM, Aspinall M . The business value and cost effectiveness of genomic medicine. Pers Med 2012;9:265–286.
Evans JP, Khoury MJ . The arrival of genomic medicine to the clinic is only the beginning of the journey. Genet Med 2013;15:268–269.
Jakka S, Rossbach M . An economic perspective on personalized medicine. The HUGO J 2013;7:1.
Hartocollis A . Cancer centers racing to map patients’ genes. New York Times 21 April 2013.
Martinez de Lecea MG, Rossbach M . Translational genomics in personalized medicine – scientific challenges en route to clinical practice. The HUGO J 2012;6:2.
Snyderman R . Personalized health care: from theory to practice. Biotechnol J 2012;7:973–979.
Centers for Medicare and Medicaid Services (CMS). Gapfill Pricing Inquiries. Issued 30 September 2013. http://www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/ClinicalLabFeeSched/Gapfill-Pricing-Inquiries.html. Accessed 16 October 2013.
Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013;15:258–267.
Sharma V, Davey JG, Humphreys C, Johnston PW . A new approach for measuring the work and quality of histopathology reporting. Histopathology 2013;63:130–136.
Hincks R . Speaking rate and information content in English linqua franca oral presentations. Engl for Specif Purp 2010;29:4–18.
Cavanagh S . Content analysis: concepts, methods and application. Nurse Res 1991;4:5–16.
Burnard P . A method of analysing interview transcripts in qualitative research. Nurse Educ Today 1991;11:461–466.
Nilsson H, Samuelsson M, Ekdahl S, Halling Y, Oster A, Perseius KI . Experiences by patients and health professionals of a multidisciplinary intervention for long-term orofacial pain. J Multidiscip Healthc 2013;6:365–371.
Reed P, Conrad DA, Hernandez SE, Watts C, Marcus-Smith M . Innovation in patient-centered care: lessons from a qualitative study of innovative health care organizations in Washington State. BMC Fam Pract 2012;13:120.
Trängeberg Sven-Olof, Ö, Stomberg MW . Listening to music during regional anesthesia: patients’ experiences and the effect on mood. J Perianesth Nurs 2013;28:291–297.
Faruki H . Genomic testing: the clinical laboratory perspective. Clin Pharmacol Ther 2013;94:190–192.
Kingsmore SF, Saunders CJ . Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Sci Transl Med 2011;3:87ps23.
Solomon BD, Nguyen AD, Bear KA, Wolfsberg TG . Clinical genomic database. Proc Natl Acad Sci USA 2013;110:9851–9855.
Deverka PA, Schully SD, Ishibe N, et al. Stakeholder assessment of the evidence for cancer genomic tests: insights from three case studies. Genet Med 2012;14:656–662.
Acknowledgements
We thank the College of American Pathologists for its support of the staff effort to conduct this study and support of the author participation through face-to-face meetings and telecommunications. We thank the survey participants for their participation and frank discussion of issues and challenges that their institutions and departments faced, and were facing, in implementing NGS testing for clinical programming. An educational webinar based on the initial study findings, sponsored by the College of American Pathologists, was presented on 11 December 2012. The recorded webinar is available at http://www.cap.org. We note the untimely passing of J.A.K. in September 2012; the loss of his contribution to the future of genomic medicine will be keenly felt.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Table S1
(DOC 38 kb)
Rights and permissions
This work is licensed. under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Crawford, J., Bry, L., Pfeifer, J. et al. The business of genomic testing: a survey of early adopters. Genet Med 16, 954–961 (2014). https://doi.org/10.1038/gim.2014.60
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2014.60
Keywords
This article is cited by
-
Cost of cancer diagnosis using next-generation sequencing targeted gene panels in routine practice: a nationwide French study
European Journal of Human Genetics (2018)