Abstract
Purpose:
Runs of homozygosity (ROHs) represent a measure of the extent of autozygosity and are correlated with the extent of inbreeding. Recently, it has been suggested that ROHs may contribute to the risk of colorectal cancer (CRC). The high rate of consanguinity and CRC in the Saudi population prompted us to test the role of autozygosity in the CRC risk.
Methods:
We compared 48 Saudi CRC patients to 100 ethnically matched controls, processed on the Affymetrix 250K StyI SNP GeneChip platform and analyzed using the plink package.
Results:
We could find no evidence of a significant relationship between autozygosity and CRC risk.
Conclusion:
The negative results in our study add additional significance to what has been previously reported in literature, as this is the first study to address these questions in an inbred population. Our subgroup analysis of patients with microsatellite unstable–positive tumors as compared with other groups did not significantly change our results. Although these results do not rule out the presence of recessively acting CRC-predisposing genes in a small percentage of patients, which our relatively small sample size could not capture, they suggest that such genes are unlikely to account for the disturbingly high incidence of CRC in our consanguineous population.
Genet Med 2012:14(8):720–728
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a major cause of mortality and morbidity worldwide. In Saudi Arabia, the incidence of CRC is increasing. According to the latest statistics, CRC is the second most common cancer among Saudi males and the third most common among Saudi females.1,2
Genetic aspects of CRC range from Mendelian forms as in familial CRC syndromes (hereditary nonpolyposis colorectal cancer and familial adenomatous polyposis) at one end of the spectrum to sporadic occurrence, which is believed to be the result of interaction of genetic and environmental factors. Although the genetic factors underlying familial CRC syndromes have been delineated, little is known about the genetic risk determinants of CRC in the general population. Current clues that suggest the involvement of recessively acting genes are based on the data associated with consanguinity and from populations that are characterized by a high degree of inbreeding3,4,5,6,7,8 as well as from studies in animals.9 The Saudi population is known for having relative genetic homogeneity due to particular demographic, historic, and tribal characteristics and is known for high consanguinity.10 The effect of inbreeding on cancer is likely more complex than simple Mendelian genetics, with many genetic components involved. Nonetheless, studying these genetically isolated populations may eventually lead to discovery of other genes that contribute to cancer predisposition.
Autozygosity is a term that is used to denote the presence of two identical haplotypes that are derived from an ancestor shared by both parents. It essentially represents a special type of homozygosity. Runs of homozygosity (ROHs) in the genome can be used to detect autozygosity and are directly correlated with the extent of inbreeding. Although the role of ROHs in unmasking recessively acting mutations is well established in Mendelian genetics, much less is known about their contribution to more complex disorders such as cancer. Assessment of ROHs on a genome-wide basis, therefore, provides a measure of extent of autozygosity and ultimately exposing recessively acting disease genes.11 Previously, a significant increase in the frequency of homozygosity in combined cases as compared with controls was reported in patients with breast, prostate, or head and neck cancer of Northern/Western European ancestry by whole-genome loss of heterozygosity analysis using microsatellite markers.12 In addition, Bacolod et al. demonstrated, using Affymetrix SNP arrays (Santa Clara, CA), that cases with CRC harbored significantly more homozygous regions than did healthy individuals.13 However, this observation could not be replicated.14,15,16 Findings from these studies support the hypothesis that there exist multiple, recessive, cancer-predisposing loci that are not readily detected using a conventional genome-wide association approach based on analysis of individual single-nucleotide polymorphisms (SNPs). Although the efforts of genome-wide association studies can help identify common variants, there are likely to be rare variants that may be uncovered through whole-genome homozygosity analysis.
To examine whether homozygosity is associated with an increased risk of developing CRC and to search for novel recessively acting disease loci, we conducted a whole-genome homozygosity analysis of 48 cases with CRC and 100 controls using the Affymetrix GeneChip Human Mapping 250K Sty Array SNP genotyping platform, asking four specific questions. First, do patients with CRC have enrichment for ROHs in particular chromosomal regions as compared with controls? Second, do patients with CRC have longer ROHs as compared with controls? Third, is there a particular SNP that is more likely to be homozygous in patients with CRC as compared with controls? And fourth, are patients with CRC more inbred than controls? To each of these questions in our study, the answer was negative, and thus we found no evidence to support the existence of an association between CRC and increased levels of homozygosity in our study population.
Materials and Methods
Patient selection and DNA extraction
Blood samples from 48 cases with CRC were provided by the Colorectal Unit, Department of Surgery, King Faisal Specialist Hospital and Research Centre with long-term follow-up data. A total of 100 matched controls were available from the Blood Bank at King Faisal Specialist Hospital and Research Centre. An on-staff pathologist (P.B.) reviewed all tumors for grade and histological subtype. The institutional review board of the King Faisal Specialist Hospital and Research Centre approved the study.
SNP array procedure
The procedure for the Affymetrix GeneChip Human Mapping 250K Sty SNP array was carried out according to the manufacturer’s guidelines. Briefly, 0.25 μg of genomic DNA was digested with StyI. The digests were then ligated to oligonucleotide adapters, polymerase chain reaction (PCR)-amplified (such that the amplicons were in the range of 250–2,000 bp), fragmented, biotin-labeled, and hybridized to the array for 16 h. Following hybridization, the array chips were washed and then stained with streptavidin–phycoerythrin and a biotinylated anti-streptavidin antibody on the Affymetrix Fluidics Station 450. The arrays were scanned in the GeneChip Scanner 3000 to generate image (DAT) and cell intensity (CEL) files. All CEL files were imported into the Affymetrix Genotyping Console analysis software 3.0 (Affymetrix) for quality control and to generate SNP calls using the Bayesian robust linear model with Mahalanobis distance classifier algorithm (http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf).
Sequencing analysis
PCR amplification of coded regions with intron–exon boundaries for selected genes (see Supplementary Table S1 online) and direct sequencing of both strands was performed. The efficiency and quality of the amplification PCR were confirmed by running PCR products on a 2% agarose gel. The PCR products were subsequently subjected to direct sequencing PCR with BigDye terminator V 3.0 cycle sequencing reagents (Applied Biosystems, Foster City, CA). The samples were finally analyzed on an ABI PRISM 3100xl Genetic Analyzer (Applied Biosystems).
Statistical methods
SNP genotyping quality control and allele-calling of the 48 CRC and 100 normal control samples were performed using the Affymetrix Genotyping Console 3.0 analysis software (http://www.affymetrix.com/products_services/software/specific/genotyping_console_software.affx). Only samples for which at least 95% of the full SNP panel had genotype calls were included in further analysis—this yielded 44 CRC and 95 normal samples. Power analysis performed for the 44 CRC and 95 control samples demonstrated that all percentages of homozygous frequencies in Table 1 for both cases (CRC) and controls (normal) are at least 30%, with odds ratio ≥3.5, suggesting that the power of the comparison in our study is close to 70% (25% when corrected for multiple testing, see Supplemental Table S2 online). For further downstream analyses of individual SNP homozygosity, inbreeding, and individual ROH, using both full and low-linkage disquilibrium (LD) SNP panels, the plink package (http://pngu.mgh.harvard.edu/purcell/plink/) was used, with default parameters except for minimum length of ROH, minimum number of SNPs per ROH, and maximum number of heterozygous SNPs per ROH. In addition, the frequency of ROHs in multiple samples was verified, along with any co-occurring copy-number variation (CNV) regions, using Genotyping Console 3.0. All statistical tests performed in this study were two-tailed. Comparison and association test statistics and plots were generated using the R Project for Statistical Computing Software (http://www.r-project.org/).
Results
To address the four questions that pertain to the relationship between autozygosity and CRC risk, we performed several tests to assess the homozygosity for SNPs and ROHs between cases and controls. We further analyzed the associations between homozygosity and CRC in subgroups of CRC. All analyses were performed using both the 250K and the low-LD SNP panels.
Genome-wide assessment of associations between homozygosity at individual SNPs and CRC risk
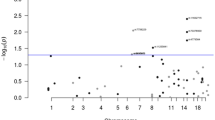
We initially tested the association between homozygosity (for either major or minor allele) and CRC risk for individual SNPs in the 250K panel. Details of the results for the seven SNPs with false-discovery rate <0.05 are shown in Table 1 ( Figure 1a ). The most strongly associated SNP was rs7936589 (chr11:23,012,445 base; P value = 7.28 × 10−7; odds ratio 9.51). No single SNP reached globally significant association (considered to be P value <10−7, using the widely accepted threshold for genome-wide association studies); Figure 1a ). To assess the correlation of genome-wide homozygosity with CRC, we aggregated homozygous SNP counts in cases versus controls, without considering the minor allele frequency. For the SNPs from the entire 250K panel, the median number of homozygous SNPs in cases was 130,613 (interquartile range (IQR) = 2,677), as compared with 130,498 (IQR = 2,930) in controls (Wilcoxon P value = 0.987). This lack of association was confirmed when we repeated the analysis using the low-LD panel, with medians of 32,782 (IQR = 695) and 32,804 (IQR = 769), respectively (P = 0.957, Wilcoxon test; Table 2 ). In addition, we calculated the inbreeding coefficient (F) across all samples (see Supplementary Tables S3 and S4 online). The median (and IQR) for F in cases and controls using all SNPs were 0.029 (0.042) and 0.023 (0.063), respectively, not significantly different from each other (Wilcoxon P value = 0.151), with similar results for the low-LD SNP panel ( Table 2 ). Therefore, we could find no evidence to suggest that cases were, in general, more inbred than controls.
SNP- and ROH-based association of CRC cases and controls. (a) SNP-based homozygosity association. (b) ROH counts by chromosomes (≥20 Mb). (c) Boxplot showing total ROH length by minimum number of SNPs between cases and controls. (d) Boxplot showing total ROH length by minimum length between cases and controls. Ctrl, control; het, heterozygote; hom, homozygote; ROH, run of homozygosity; SNP, single-nucleotide polymorphism.
Analysis of ROHs in cases and controls
The existence of LD blocks means that relatively short ROHs, from tens to hundreds of kilobases, are common across the genome.17 Evidently, most of these regions probably do not result from true autozygosity. We therefore set thresholds to define an ROH based on genomic regions where either a minimum number of (50) consecutive, nonmissing SNPs were homozygous (allowing for miscalls) or in which homozygosity extended for a minimum length (250 kb) along the chromosome. We also calculated total ROH length per individual (i.e., the sum of the lengths of the ROHs in their genome) as a more robust measure of autozygosity than counting the number of ROHs per genome. For example, with a threshold ROH size of 4 Mb, the latter method would twice score two ROHs of 4 Mb but would only score a single region of 8 Mb once; in terms of indicating autozygosity, however, a single 8-Mb region would be at least as important as two 4-Mb regions.
To provide a comparison with the work of Bacolod and colleagues,13 we initially analyzed ROHs that were ≥50 SNPs in size using the 250K SNP panel. Every individual had at least 22 ROHs and the median number of these regions per individual was approximately 148. There was no evidence of an association between the total ROH size in each individual and CRC (P = 0.201, Wilcoxon test; Table 3 ). To determine whether this result was robust, we repeated the analysis using a number of different criteria to define a ROH (≥30 SNPs, ≥40 SNPs, ≥60 SNPs, ≥1 Mb, ≥2 Mb, ≥4 Mb, and ≥10 Mb; Figure 1b–d ). The only evidence found for an association between total size of the ROHs in each individual and CRC was for ROHs containing ≥30 SNPs ( Table 3 ). However, this result was not confirmed on repeating the analysis in the low-LD SNP panel using the same ROH defining criteria as above. Every individual carried at least one homozygous region (median 4.0) detected by the low-LD SNP panel and virtually all of these regions were large (1–42 Mb in size). Analysis of the total size of ROHs in each individual determined again that there was no significant difference between cases and controls ( Table 3 ). The total length of ROHs detected in each person using the 250K and low-LD SNP panel are summarized in ( Figure 1c,d ) and detailed in Supplementary Tables S3 and S4 online, respectively. Figure 1b illustrates the overall distribution of large ROHs (≥20MB) across all chromosomes.
To provide a further comparison between our results and those of Bacolod and colleagues,13 we calculated the frequencies of cases and controls in which we detected one or more ROHs of >4 Mb in length. Using the 250K SNP panel, 39 of 44 (88.6%) cases and 72 of 95 (75.8%) controls had these ROHs (P = 0.11, Fisher’s exact test). For the low-LD panel, 29 of 44 (65.9%) cases and 47 of 95 (49.5%) controls had ROHs of >4 Mb (P = 0.10, Fisher’s exact test). We thus failed to detect the marked significant difference between cases (62.2%) and two sets of controls (35.6% and 28.8%) that was seen in the study by Bacolod and colleagues. The longest homozygous regions were derived from individuals in our study who were found to have higher levels of inbreeding (F >0.06) than the median (F = 0.026 for all samples) from a population with strong evidence of consanguineous marriage.
Recurrent ROHs
Although most of the ROHs examined thus far were individually uncommon, some occurred in >10% of cases or controls when assessed using the 250K panel. We therefore addressed whether any of these specific, relatively common homozygous regions were associated with CRC risk. Using ROHs with ≥1 Mb of consecutive homozygous SNPs detected in the 250K panel, we searched for overlapping ROHs that were found in more than five individuals (cases and/or controls). This resulted in a total of 4,169 ROHs that met the inclusion criteria. After taking multiple testing into account, 12 ROHs reached global significance for an association between homozygosity and CRC risk ( Table 4 ), all of which were more common in cases than controls. One ROH was found in five different samples at chromosome 11q near the centromere.
Confirmation of homozygous regions
Homozygous regions might result from chance, autozygosity, uniparental isodisomy, or hemizygosity. To determine whether the common homozygous regions were actually hemizygous CNVs, the positions of the common ROHs were compared with those of CNVs identified using Genotyping Console 3.0. We searched for CNVs that covered at least 90% of the detected ROH. However, none of the common ROHs we found could be explained by a CNV (data not shown).
Additionally, a subgroup of patients with microsatellite unstable (microsatellite unstable–positive) tumors were compared with the negative group. Molecular diagnosis showed that 7 of 48 cases were microsatellite unstable–positive. After correcting for multiple testing, eight ROHs reached global significance for an association between homozygosity and CRC risk in microsatellite unstable–positive cases, which were shared by five of the six samples. For selected genes from these regions, the encompassing coding region with intron–exon boundaries were sequenced; however, no mutations were detected (see Supplementary Table S5 online). Analysis with subgroups did not significantly change our results with respect to our four hypothesized questions (data not shown).
Discussion
The most plausible explanation for the presence of long stretches of homozygous regions in an individual’s genome is that his or her parents can trace their lineage to a common ancestor. That these regions resulted from uniparental disomy (an instance when an offspring inherits both copies or segments of chromosomes from a single parent), although possible, is highly unlikely. Recent studies12,13 have reported an increased frequency of homozygous microsatellites or ROHs in cancer cases as compared with controls and that these regions showing identity by descent may be the locations of genes contributing to tumor heritability.12,13 Moreover, these data have been interpreted as providing an explanation for the increased cancer rates in populations with higher degrees of consanguinity. There are studies that have compared the incidence of cancer and other late-onset complex diseases between individuals from genetically isolated islands in Middle Dalmatia, Croatia, and a control population.18 These studies suggest that inbreeding can be a positive predictor for a number of late-onset diseases such as heart disease, stroke, and cancer. Similar observations were published in a Pakistani study where cancer patients, on average, have a higher coefficient of inbreeding as compared with the general population.7 Another study demonstrated that 94% of the subjects with reported adenocarcinomas (mostly colorectal) originated from a consanguineous population of descendants of an Italian immigrant group in Wisconsin,6 suggesting an explanation for increased cancer incidence with higher degree of inbreeding. Thus, an explanation for increased cancer risk based on the frequency of homozygous regions and consanguinity has formed the basis of a potentially new model of cancer progression.19
In this study, we have used Affymetrix GeneChip 250K SNP arrays to compare the structure of genetic variation in patients with CRC to that of healthy controls. Our analyses analyzed SNPs from both the full panel as well as those in low pairwise LD. Overall, evidence for an association between homozygosity and CRC risk was limited. Our results demonstrated a total of 4,169 ROHs that were found in more than five individuals (either cases or controls) and met the inclusion criteria. After taking multiple testing into account, only 12 ROHs reached significance for an association between homozygosity and CRC risk ( Table 4 ), all of which were more common in cases than controls. Sequencing of selected genes with regions, including coding and intron/exon boundaries yielded no mutations (see Supplementary Table S1 online). We did not find cases to be significantly more or less inbred than controls. Furthermore, our ROH analysis provided no evidence for an association between total ROH size per individual and increased risk of CRC, under any of several size criteria, using either the 250K or the low-LD SNP panel.
The assertion that increased autozygosity correlates with cancer incidence provides an attractive explanation for reported increased cancer risk in inbred populations. However, several criticisms can be leveled at this assertion. The observation of an increased cancer risk associated with consanguinity has often been based on studies of a small number of individuals in an isolated community or a single large family with a high level of inbreeding.20 Thus, the relevance of inbreeding to the population risk of cancer is unclear, as inbreeding and founder effects may be confounded. Lack of confirmation of these results by our study could partially be attributable to sample size, which was relatively small. Nonetheless, the negative results in this study of matched cases and controls from an inbred population carry more significance than what has been previously reported in the literature. Previous molecular studies have sought to establish a relationship between ROHs and cancer risk with case–control groups that have been ethnically heterogeneous or unmatched.12,13 In addition these studies made use of relatively sparse microsatellite data, whereby, in our small cohort case–control study of CRC we have addressed these shortcomings by analyzing samples using a genome-wide 250K SNP platform and imposed a high level of quality control both in terms of genotyping and sample ancestry. Furthermore, our data from a subgroup of patients with microsatellite unstable–positive tumors did not significantly change our results. Of note, our sample set did not identify homozygosity across MYH, perhaps the only known recessively acting CRC gene. It is possible that our study sample size is not large enough to detect this or similarly recessive genes.
Our results are concordant with similar studies carried out in different cancers, such as breast, prostate, leukemia, and colon cancer,14,15,16 in a predominantly consanguineous population. Because our analysis suggests that whole-genome homozygosity analysis of inbred populations may not provide a robust methodology for identifying novel cancer susceptibility loci, it would be worthwhile to apply new strategies such as exome or whole-genome sequencing in the future to unveil these underlying, predisposing recessive alleles. It is unlikely that large numbers of recessive alleles exist that predispose to CRC and would be unmasked by autozygosity in inbred populations, such as that in Saudi Arabia.
In conclusion, our findings do not provide evidence that increased levels of homozygosity confer an increased risk of developing CRC. Although these results do not rule out the potential presence of recessively acting CRC-predisposing genes in a small percentage of patients that our relatively small sample size could not capture, they do suggest that such genes are unlikely to account for the disturbingly high incidence of CRC in our consanguineous population and that future research should consider other mechanisms.
Disclosure
The authors declare no conflict of interest.
References
Bavi PP, Abubaker JA, Jehan ZD, et al. Colorectal carcinomas from Middle East. Molecular and tissue microarray analysis of genomic instability pathways. Saudi Med J 2008;29:75–80.
Al-Eid HS . Cancer Incidence Report Saudi Arabia 1999–2000. King Faisal Specialist Hospital and Research Centre, 2004. http://www.scr.org.sa/reports/SCR2000.pdf.
Bener A, El Ayoubi HR, Chouchane L, et al. Impact of consanguinity on cancer in a highly endogamous population. Asian Pac J Cancer Prev 2009;10:35–40.
Feldman JG, Lee SL, Seligman B . Occurrence of acute leukemia in females in a genetically isolated population. Cancer 1976;38:2548–2550.
Abramson JH, Pridan H, Sacks MI, Avitzour M, Peritz E . A case-control study of Hodgkin’s disease in Israel. J Natl Cancer Inst 1978;61:307–314.
Lebel RR, Gallagher WB . Wisconsin consanguinity studies. II: Familial adenocarcinomatosis. Am J Med Genet 1989;33:1–6.
Shami SA, Qaisar R, Bittles AH . Consanguinity and adult morbidity in Pakistan. Lancet 1991;338:954.
Simpson JL, Martin AO, Elias S, Sarto GE, Dunn JK . Cancers of the breast and female genital system: search for recessive genetic factors through analysis of human isolate. Am J Obstet Gynecol 1981;141:629–636.
Demant P . Cancer susceptibility in the mouse: genetics, biology and implications for human cancer. Nat Rev Genet 2003;4:721–734.
el-Hazmi MA, al-Swailem AR, Warsy AS, al-Swailem AM, Sulaimani R, al-Meshari AA . Consanguinity among the Saudi Arabian population. J Med Genet 1995;32:623–626.
Alkuraya FS . Autozygome decoded. Genet Med 2010;12:765–771.
Assié G, LaFramboise T, Platzer P, Eng C . Frequency of germline genomic homozygosity associated with cancer cases. JAMA 2008;299: 1437–1445.
Bacolod MD, Schemmann GS, Wang S, et al. The signatures of autozygosity among patients with colorectal cancer. Cancer Res 2008;68:2610–2621.
Enciso-Mora V, Hosking FJ, Houlston RS . Risk of breast and prostate cancer is not associated with increased homozygosity in outbred populations. Eur J Hum Genet 2010;18:909–914.
Hosking FJ, Papaemmanuil E, Sheridan E, et al. Genome-wide homozygosity signatures and childhood acute lymphoblastic leukemia risk. Blood 2010;115:4472–4477.
Spain SL, Cazier JB, Houlston R, Carvajal-Carmona L, Tomlinson I; CORGI Consortium,. Colorectal cancer risk is not associated with increased levels of homozygosity in a population from the United Kingdom. Cancer Res 2009;69:7422–7429.
Abecasis GR, Ghosh D, Nichols TE . Linkage disequilibrium: ancient history drives the new genetics. Hum Hered 2005;59:118–124.
Rudan I, Rudan D, Campbell H, et al. Inbreeding and risk of late onset complex disease. J Med Genet 2003; 40:925–932.
Bacolod MD, Schemmann GS, Giardina SF, Paty P, Notterman DA, Barany F . Emerging paradigms in cancer genetics: some important findings from high-density single nucleotide polymorphism array studies. Cancer Res 2009;69:723–727.
Akasaka T, Balasas T, Russell LJ, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood 2007;109:3451–3461.
Acknowledgements
This work was supported by grants from the King Abdulaziz City for Science and Technology (KACST) and the National Comprehensive Plan for Science and Technology resulting from KACST Project 08-MED479-20.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Table S1.
(DOC 45 kb)
Supplementary Table S2.
(DOC 71 kb)
Supplementary Table S3.
(DOC 1067 kb)
Supplementary Table S4.
(DOC 884 kb)
Supplementary Table S5.
(DOC 32 kb)
Rights and permissions
About this article
Cite this article
Siraj, A., Khalak, H., Sultana, M. et al. Colorectal cancer risk is not associated with increased levels of homozygosity in Saudi Arabia. Genet Med 14, 720–728 (2012). https://doi.org/10.1038/gim.2012.27
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2012.27
Keywords
This article is cited by
-
Runs of homozygosity: windows into population history and trait architecture
Nature Reviews Genetics (2018)
-
Breast cancer protection by genomic imprinting in close kin families
BMC Medical Genetics (2017)
-
Runs of homozygosity and inbreeding in thyroid cancer
BMC Cancer (2016)
-
Impact of Physiological Symptoms and Complications of Colorectal Cancer on the Quality of Life of Patients at King Abdulaziz University Hospital
Journal of Cancer Education (2016)