Abstract
Purpose: A population-based cohort from three state newborn screening programs was used to describe β-globin gene cluster variation.
Methods: Blood spots from newborns homozygous for HbS were genotyped for five restriction fragment length polymorphisms (RFLPs) to construct β-globin haplotypes. Haplotype distributions were compared by race/ethnicity and sex. Expected heterozygosities were calculated and compared with observed heterozygosities.
Results: Haplotype distributions did not differ between sexes for either blacks or Hispanics. Neither racial/ethnic group deviated from Hardy-Weinberg equilibrium; however, Hispanics had higher heterozygosity at two RFLPs compared with blacks.
Conclusion: The differences between populations probably reflect recent migration and admixture rather than selection.
Similar content being viewed by others
Main
Sickle cell disease refers to a group of autosomal recessive hemoglobinopathies involving the β-globin gene cluster on chromosome 11 that is manifested by the presence of sickle hemoglobin (HbS) in red blood cells. In the United States, sickle cell disease is one of the most common genetic disorders, with a carrier frequency of 1 in 14 blacks, and it causes considerable morbidity and mortality in the general population.1–3 The most common form of sickle cell disease is often referred to as sickle cell anemia, which is the hemoglobinopathy caused by the inheritance of two copies of a single mutation in the β-globin gene: glutamic acid to valine in codon 6.
With the advent of recombinant DNA technology, in addition to its clinical importance, the study of the HbS mutation within the β-globin gene cluster has contributed scientific knowledge to broader areas of population genetics. First, studies of the origin of the HbS mutation were important in understanding the mutational process in the human genome. Early studies of genetic variation within the β-globin gene cluster demonstrated that the HbS mutation was found on only a few haplotype backgrounds, suggesting multiple origins of the HbS mutation.4–7 These haplotype backgrounds are found where the HbS mutation presumably originated in human history: Benin (BEN), Cameroon (CAM), Central African Republic (CAR), Senegal (SEN), and Saudi Arabia (SAUDI). Alternatively, these multiple haplotype backgrounds with the HbS mutation may represent gene conversion events occurring in genetically isolated populations.8 Archaeological and genetic data indicate that the HbS mutation is evolutionarily recent, with estimates that the mutation first occurred <3,000 to 10,000 years ago.8,9
Second, the β-globin gene cluster and the HbS mutation are important for understanding the selective forces that shape the distribution of genetic variation within and among human populations. While persons with homozygous HbS have life-threatening complications, early observational studies have suggested that persons with sickle cell trait (heterozygous HbS) are protected from Plasmodium falciparum malaria.10,11 This protective factor makes the β-globin gene cluster an appropriate area to examine for the genetic signatures of natural selection and genetic drift within human populations.8 Also, the recombination hot spot in the 3′ region of the β-globin gene cluster makes this region of the genome important for studies of how human recombination impacts genetic variation.12 Finally, the genetic variability and linkage disequilibrium around the β-globin gene cluster, coupled with the high prevalence of the HbS mutation in many areas of the world, make the β-globin gene cluster particularly useful for studies of human evolution.13–18
In addition to its contribution to population genetics, the study of the β-globin gene cluster has also proven valuable in the study of disorders that are modified by other genes. Although sickle cell anemia (HbS) is a single-gene disorder, clinical variability among those with the same genotype has been documented extensively.19 Studies of the determinants for this clinical variability suggest that several trans-acting factors, such as coinheritance of α-thalassemia, may be involved.20Cis-acting factors, including haplotype background, have also been implicated in determining clinical variability among persons homozygous for the HbS mutation.21–23
While β-globin gene cluster variation has been characterized for more than 20 years in various populations, very little information has been gathered on the distribution of this variation in persons with homozygous HbS using a population-based study design. Many population studies that characterize β-globin haplotypes in the United States were small and/or were drawn from a single state.4,7,24–29 Also, most population and clinical studies involving the β-globin gene in the United States describe adults ascertained in a clinic setting. This type of ascertainment can introduce biases in studies examining the relationship between clinical severity and haplotype background if a subset of children with a specific genetic background does not survive into adulthood.
In an effort to collect population-based data on trends and risk factors from persons with homozygous HbS born in the United States, the Centers for Disease Control and Prevention (CDC) funded follow-up studies of newborns with sickle cell disease through the universal newborn screening programs in California, Illinois, and New York.30 During the study period, 1992 through 1993, the three states included in this study represented approximately 20% of all infants born to blacks in the United States.31 We report here the only population-based study for β-globin gene haplotypes in the United States for persons with homozygous HbS ascertained in a nonclinic setting. These results provide the groundwork for studies determining the contribution of haplotype background to clinical severity. These results also provide an opportunity to describe admixture in the United States with respect to the HbS mutation.
METHODS
Study population
Children with sickle cell disease (N = 1,042) were identified through newborn screening programs in California (n = 265), Illinois (n = 254), and New York (n = 523) between January 1, 1992, and December 31, 1993.30 A family survey designed to collect demographic variables and risk factors for complications was administered by telephone. The survey was intended for families of all children in the cohort; however, many families could not be located and only a small proportion (n = 252; 24%) could be contacted for the survey. In addition to the family survey, a questionnaire designed for physicians was mailed to the child’s last known hematologist or health care provider. A larger proportion of physician questionnaires were returned (n = 752; 72%). This study was approved by the institutional review boards of the CDC and the individual state health departments of California, Illinois, and New York.
Genotyping methods
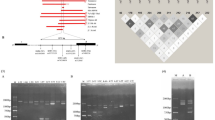
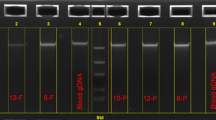
Specimens were coded by personnel at each state site and delivered to the Wadsworth Center, Biggs Laboratory. DNA extraction was carried out as previously described.32 Specimens were punched with a 1-mm hand-held punch that was sterilized with ethanol flame between punches. One hundred microliters of methanol was added to each sample, and the specimens were allowed to air-dry overnight in a fume hood. After fixation, samples were hydrated in 25 μL of sterile, distilled water and placed into a boiling water bath for 10 minutes. Polymerase chain reaction (PCR) racks were cooled to room temperature and centrifuged for 3 minutes at 1,200 ×g before PCR setup. Four of the five restriction fragment length polymorphisms (RFLPs) were detected in two multiplex reactions. The fifth RFLP was detected as a single reaction. Polymorphisms at the Xmn I restriction site in the Gγ region and the Hin fI RFLP 5′ of the β-globin gene were multiplexed using the following PCR primers: SC0A 5′AAA ATT GGA ATG ACT GAA TCG GAA CAA GGC AAA G3′; SC1A 5′TAT TGA TAA CCT CAG ACG TTC CAG AAG CGA GTG TG3′ (308 bp for Gγ); SC9X 5′GCG GTC CCA AAA GGG TCA GTC TAC GCT GAC CTC ATA AAT G3′; SC10X 5′GCG GTC CCA AAA GGG TCA GTC TAA TCT GCA AGA GTG TCT3′ (424 bp for 5′β). Polymorphisms at the Hin cII sites in the φβ region and the 5′δ region were also multiplexed using the following PCR primers: SC5X 5′5′GCG GTC CCA AAA GGG TCA GTG AAC AGA AGT TGA GAT AGA GA3′; SC6X 5′GCG GTC CCA AAA GGG TCA GTA CTC AGT GGT CTT GTG GGC T3′ (700 bp for φβ); SC7X 5′GCG GTC CCA AAA GGG TCA GTT CTG CAT TTG ACT CTG TTA GC3′; SC8X 5′GCG GTC CCA AAA GGG TCA GTG GAC CCT AAC TGA TAT AAC TA3′ (613 bp for 5′δ). The Hin dIII polymorphism in the Gγ region also was amplified using SC2 5′AAG TGT GGA GTG TGC ACA TGA3′ and SC3 5′TGC TGC TAA TGC TTC ATT ACA A3′ (781 bp). Amplification for the Xmn I/ Hin fI RFLPs were carried out in a 50-μL volume containing 10 mM Tris-HCl, pH 8.8; 50 mM KCl; 200 μM each dATP, dTTP, dCTP, and dGTP; 2.5 mM MgCl2; and 2.5 U Taq DNA polymerase, 16:1 molar ratio Taq Start Antibody (Clontech, Palo Alto, CA):Taq polymerase, 0.5 μM each primer, and 25 μL of the DNA extract. Conditions for the other polymorphisms were the same except that 2.0 mM MgCl2 was used in the Hin cII and Hin dIII reactions. Finally, buffer for the Hin dIII reaction was pH 8.3. All samples were heat denatured at 95°C for 5 minutes followed by 35 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds (PE9600). A final elongation step was carried out for 5 minutes at 72°C.
Samples were also genotyped to identify the presence of the HbS allele in both chromosomes using primers TB1 5′GCG GTC CCA AAA GGG TCA GTT CCT AAG CCA GTG CCA GAA G3′and PC04 5′GCG GTC CCA AAA GGG TCA GTC AAC TTC ATC CAC GTT CAC C3′. Amplification was carried out in a 50-μL volume containing 10 mM Tris-HCl, pH 8.3; 50 mM KCl; 200 μM each dATP, dTTP, dCTP, and dGTP; 1.5 mM MgCl2; 0.8 U Taq DNA polymerase, 16:1 molar ratio Taq Start Antibody (Clontech, Palo Alto, CA):Taq polymerase; 0.1 μM each primer; and 25 μL of the DNA extract using the same thermal cycler and program as above. Digestions were carried out for each polymorphism:Xmn I (6 U)/ Hin fI (10 U), Hin cII (10 U), Hin dIII (20 U), and Dde I (2.4 U) for the HbS typing per manufacturer’s recommendations. All enzymes were purchased from New England Biolabs (Beverly, MA) except Hin cII (Promega Corporation, Madison, WI). Digestion products were electrophoresed on a 2% NuSieve:1% LE agarose gel (BioWhittaker, Inc., Rockland, ME) and made visible by the use of ethidium bromide staining.
Statistical methods
Allele frequencies at individual RFLPs were estimated by gene counting. Haplotype distributions were compared using the software StatXact 3.0 (Cytel Software Corporation, Cambridge, MA) or Statistical Analysis Battery for Epidemiologic Research (SABER) version 1.96.33 Specifically, the distribution of haplotypes identified in this cohort was examined in two ways. First, the number of haplotypes among chromosomes was determined in each population. Because each person has a maternal and paternal chromosome,11 each person will have two haplotypes counted in this analysis. Second, the number of haplotype combinations identified within each person was determined. In this count, only one haplotype combination can be identified per person. These approaches to describing the haplotype distribution are analogous to examining the allele distribution (p and q) and the genotype distribution (p,2 2pq, q2), respectively, of a biallelic marker in the same population. We tested for possible differences between distributions by a χ2 test or Fisher exact, as indicated in the text. Haplotypes other than BEN, CAM, CAR, SEN, and SAUDI were considered minor and classified as “Other” for allele and haplotype distribution comparisons. Expected heterozygosities (H) were estimated as n (1 − Σpi2)/(n − 1), where pi represents the frequency of the i th allele or haplotype and n is the sample size. Standard errors were calculated as [(1 − H)H/ n],1/2 where H is the expected heterozygosity and n is the sample size. Differences between observed and expected heterozygosities were determined by performing a χ2 test. Genotypes that could not be ordered for haplotypes were excluded from haplotype heterozygosity counts and calculations, but were included as “Other” for simple haplotype counts in Tables 1 and 2.
RESULTS
Among the 1,042 newborns identified with sickle cell disease through newborn screening programs, 617 (60%) were determined to be homozygous (HbSS) for the sickle hemoglobin mutation. Stored blood spots were genotyped successfully for the markers used to determine β-globin haplotypes for 430 (70%) of the newborns with HbSS, with 244 (57%) from New York, 124 (29%) from California, and 62 (14%) from Illinois. Of the 187 samples that failed to yield genotype information, 90 (48%) were from Illinois, 81 (43%) were from New York, and 16 (9%) were from California.
To determine race/ethnicity of the 430 newborns with genotype information, responses to the race/ethnic background question in the parental interview were used. For newborns identified with sickle cell disease whose parents did not complete a parental survey, race/ethnicity was abstracted from newborn screening program demographic information supplemented by additional information from the sickle cell disease registry in California and pediatric hematologists in Illinois. Most newborns with genotype information were black, not of Hispanic origin (n = 371; 86.3%) and Hispanic (n = 32; 7.4%). The remaining newborns were reported to be of mixed race/ethnicity or Other (n = 7), Asian or Pacific Islander (n = 1), and white, not of Hispanic origin (n = 1). We were unable to determine race/ethnicity for 18 (4.2%) newborns with haplotype information. Of the 187 samples that did not yield genotype information, 164 had information on race/ethnicity of the newborn. The distribution of race/ethnicity of the samples that failed to yield genotype information did not differ from the samples with haplotype information (data not shown).
β-Globin haplotypes were determined by genotyping five RFLPs within the β-globin gene cluster (Fig. 1). The haplotype distribution did not differ among chromosomes from the three different states (data not shown); therefore, all three states were combined for subsequent analyses. Among chromosomes from the black population (n = 742), the BEN haplotype was the most frequent (63%), followed by the CAR and SEN haplotypes (14% and 9%, respectively;Table 1). Some chromosomes from the black population (6%) had minor haplotypes (n = 38) or genotypes whose order could not be determined (n = 6). The distribution of haplotypes among chromosomes from black females did not differ from the distribution among chromosomes from black males (χ25 = 8.16, P > 0.05;Table 1). Also, within the black population, the overall distribution of haplotype combinations did not differ when stratified by sex (Fisher exact, P > 0.05;Table 2). In total, at least 16 distinct haplotypes were identified among chromosomes, and 28 haplotype combinations were identified among the black population (Tables 1, 2, and 3).
Compared with the chromosomes from blacks, chromosomes from the Hispanic population (n = 64) had a slightly different distribution of β-globin haplotypes. The BEN haplotype was the most common haplotype among chromosomes from the Hispanic population, as was observed among chromosomes from the black population. The proportion of CAR haplotypes and CAM haplotypes was higher among the Hispanic population than the black population (28% and 8%, respectively), and the proportion of SEN haplotypes was lower (6%) in the Hispanic population (Table 1). However, these differences were not statistically significant (Fisher exact, P > 0.05). Unlike the black population, the distribution of haplotypes among chromosomes from the Hispanic population differed slightly by sex, with a higher frequency of the BEN haplotype and a lower frequency of the CAR haplotype among chromosomes from Hispanic females when compared with chromosomes from Hispanic males (Fisher exact, P = 0.04;Table 1). Examination of a larger cohort of Hispanics will be needed to determine whether this difference is real or a consequence of a small sample size. Although Hispanic females had a higher frequency of homozygous BEN haplotypes and no homozygous CAR haplotypes compared with Hispanic males, the overall distribution of haplotype combinations between males and females did not differ (Fisher exact, P > 0.05;Table 2). In total, six distinct haplotypes were identified among chromosomes (Table 1) and 10 haplotype combinations were identified among the Hispanic population (Tables 2 and 3). All haplotypes identified among chromosomes from the Hispanic population were also identified among chromosomes from the black population, suggesting that the haplotypes identified among Hispanics in this study population are of African origin.
Among other racial/ethnic groups, the following haplotype combinations were identified: BEN/BEN (Asian or Pacific Islander, n = 1; Other, n = 1), BEN/CAM (Other, n = 1), BEN/CAR (Other, n = 2), BEN/Other (Caucasian, n = 1), CAR/CAR (Other, n = 2), and CAR/SEN (Other, n = 1). For newborns whose race/ethnicity could not be determined, the BEN/BEN (n = 7), BEN/CAM (n = 1), BEN/CAR (n = 3), BEN/Other (n = 1), BEN/SEN (n = 1), CAM/Other (n = 1), CAR/CAR (n = 3), and SEN/Other (n = 1) haplotype combinations were identified. All of the identified haplotypes, including the minor haplotypes, were also found among chromosomes from the black population. For the entire cohort, the following haplotype combinations were not identified in any racial/ethnic group: CAM/CAM, SEN/SEN, SEN/SAUDI, and SAUDI/SAUDI. It is interesting that the minor haplotype −, +, +, +, + found among blacks is paired exclusively with the SEN haplotype, which is one RFLP (Xmn I) shy of the homozygous SEN/SEN haplotype (Table 3).
A recent study of the β-globin gene cluster variation demonstrated that the balancing selection experienced in this region does not disturb the Hardy-Weinberg equilibrium pattern.9 Therefore, to further describe β-globin gene cluster variation in our cohort, we determined the observed heterozygosity of the individual RFLPs and haplotypes and compared these data with the calculated expected heterozygosities (see “Statistical methods”). For both blacks and Hispanics, the observed heterozygosity at each of the five RFLPs did not differ from the expected heterozygosity (Table 4). Similarly, the observed haplotype heterozygosity for blacks (0.527) and Hispanics (0.563) did not differ from the calculated expected heterozygosities (0.574, SE = 0.026 and 0.582, SE = 0.062, respectively). The Hispanic population had a higher observed heterozygosity at two of the five RFLPs, Hin dIII Gγ and Hin cII δ, when compared with the black population (Table 4). Also, the Hispanic population had a slightly higher observed haplotype heterozygosity or higher rate of haplotype combinations (0.563) compared with the black population (0.527).
DISCUSSION
We present here the only population-based description of β-globin gene cluster haplotypes in the United States. The estimated frequency of β-globin haplotypes among chromosomes from the black population is similar to that reported in a recent review of 456 chromosomes from blacks surveyed in the United States.34 To our knowledge, the estimated frequency of β-globin haplotypes among chromosomes from Hispanics is the only available description for Hispanics born in the United States. Compared with available estimates of Central/South American and Caribbean β-globin haplotype diversity, the distribution of haplotypes in this Hispanic population is similar to that described for Venezuela35 and Cuba,36 but different from that described for Jamaica,6,7,34 Guadeloupe,37 Colombia,38 and Brazil.39
As expected, the black population reported here had more distinct haplotypes and haplotype combinations than the Hispanic population. However, the Hispanic population reported here demonstrated a higher RFLP heterozygosity at Hin dIII Gγ and Hin cII δ and a higher overall haplotype heterozygosity than the black population (Table 4). This result was unexpected because population surveys of other nuclear RFLPs demonstrate that African-derived populations typically have higher levels of diversity than non-African populations.40–42 Natural selection is presumed to disrupt this pattern observed among populations, as suggested at the phenylalanine hydroxylase (PAH) locus43 and at the aldehyde dehydrogenase 2 (ALDH2) locus.44
A second force that could disrupt the typical pattern of diversity across populations is admixture. Indeed, the population of Hispanics born in the United States is a mixture of many nationalities, including persons from Mexico, Puerto Rico, Central America, South America, and the Caribbean. Also, the contribution of each nation is rapidly changing as the profile of immigrants living in the United States evolves.45 Estimates of African contribution to the Hispanic population differ depending on the origin of the population. For example, the contemporary gene pool for Mexicans is estimated to be 8% African, 31% Native American, and 61% Spanish-derived, whereas the corresponding estimates for Puerto Ricans are 37%, 18%, and 45% and for Cubans, 20%, 18%, and 62%, respectively.46 Although the origin of Hispanic populations differs among states (e.g., predominantly Mexican in California and Puerto Rican in New York47), the demographic information available for this study did not explore the nation of origin of the Hispanic population reported here. The higher diversity observed in this Hispanic population compared with this black population may be caused by admixture or the inappropriate grouping of persons of unknown Hispanic origin.
The main strength of these results is that they represent an analysis of an unselected, population-based cohort. The advantage of this study design is that it uses data that represent an unbiased sample of β-globin gene cluster variation in the three states studied. This feat was achieved through the use of universal newborn screening programs established to identify newborns affected by hemoglobinopathies.2 Also, this study was possible because the blood spots used for the initial screening were stored and could be retrieved for further genotyping of the β-globin gene cluster.48
Stored blood spots allow investigators to perform large, population-based genetic studies for specific loci; however, they also have disadvantages. One potential disadvantage for some studies is that the prevalence and distribution of genotypes obtained from the samples is derived from a cohort of newborns, which may not necessarily be identical with the prevalence and distribution derived from a cohort of adults. For the HbS mutation, the distribution of β-globin cluster haplotypes differed between children and adults in a Cuban population,36 suggesting newborns with haplotypes associated with severe complications may not live to adulthood. Although the data from that study have not been replicated,35,37,38 investigators should note that the prevalence of genotypes determined from blood spots could be limited to a newborn population, depending on the disorder being studied.
A second potential problem related to the use of blood spots is that each spot represents a diploid genome. For large-scale genetic studies involving several loci, genotypes must be ordered to construct haplotypes. In this study, a small proportion (1.4%) of the genotypes could not be ordered through inference and were discarded from subsequent analyses. Typically, ambiguous genotypes are ordered by ascertaining and genotyping several generations in a pedigree. Alternatively, molecular techniques, such as somatic cell hybrids49 and allele-specific polymerase chain reaction (AS-PCR),50 can be used to order genotypes. Both the pedigree approach and these alternative molecular approaches are expensive and time-consuming for studies with large sample sizes. Also, alternative molecular approaches may not be compatible with the type of sample obtained for the study. For the pedigree approach, ascertainment of additional family members of newborns for a population-based study may be difficult because families are often lost to tracking or are unwilling to participate in follow-up studies. Thus the use of stored blood spots from newborns may be limited to certain studies.
Finally, the extent to which blood spots may be stored and used for research is not yet clear. Many ethical, legal, and social issues surrounding the use of dried blood spots in a research setting, including linkage of the spots to personal information, are still being debated and are not fully resolved.51 While the debate continues over the research uses, analysis of stored blood spots has been useful to address issues of public health importance. For example, investigators have used stored blood spots to test new technologies for newborn screening such as tandem mass spectrometry52,53 and to determine population-based genotype frequencies of loci of interest.54–56 The need for this type of repository will not diminish because technology is constantly evolving.
In conclusion, these data provide an unbiased estimate of β-globin haplotype diversity in the United States. The data also highlight the utility of stored blood spots for large, population-based genetic studies. Although participation rates and tracking hinder follow-up studies, these data coupled with clinical outcomes could enlighten the role β-globin haplotype diversity plays in predicting clinical severity among persons with homozygous HbS.
References
Centers for Disease Control and Prevention. Mortality among children with sickle cell disease identified by newborn screening during 1990–1994—California, Illinois, New York. MMWR Morb Mortal Wkly Rep 1998; 47: 169–172.
Ashley-Koch A, Yang Q, Olney RS . Sickle hemoglobin (HbS) allele sickle cell disease: a HuGE review. Am J Epidemiol 2000; 151: 839–845.
Ashley-Koch A, Murphy CC, Khoury MJ, Boyle CA . Contribution of sickle cell disease to the occurrence of developmental disabilities: a population-based study. Genet Med 2001; 3: 181–186.
Kan YW, Dozy AM . Evolution of the hemoglobin S C genes in world populations. Science 1980; 209: 388–391.
Antonarakis SE, Boehm CD, Giardina PJ, Kazazian HH . Nonrandom association of polymorphic restriction sites in the beta-globin gene cluster. Proc Natl Acad Sci U S A 1982; 79: 137–141.
Wainscoat JS, Bell JI, Thein SL, Higgs DR, Sarjeant GR, Peto TE, Weatherall DJ . Multiple origins of the sickle mutation: evidence from beta S globin gene cluster polymorphisms. Mol Biol Med 1983; 1: 191–197.
Antonarakis SE, Boehm CD, Serjeant GR, Theisen CE, Dover GJ, Kazazian HH . Origin of the beta S-globin gene in blacks: the contribution of recurrent mutation or gene conversion or both. Proc Natl Acad Sci U S A 1984; 81: 853–856.
Flint J, Harding RM, Clegg JB, Boyce AJ . Why are some genetic diseases common? Distinguishing selection from other processes by molecular analysis of globin gene variants. Hum Genet 1993; 91: 91–117.
Currat M, Trabuchet G, Rees D, Perrin P, Harding RM, Clegg JB, Langaney A, Excoffier L . Molecular analysis of the beta-globin gene cluster in the Niokholo Mandenka population reveals a recent origin of the beta(S) Senegal mutation. Am J Hum Genet 2002; 70: 207–223.
Raper HB . Sickling in relation to morbidity from malaria other diseases. BMJ 1956; 1: 965–966.
Allison AC . Polymorphism natural selection in human populations. Cold Spring Harb Symp Quant Biol 1964; 29: 137–149.
Schneider JA, Peto TA, Boone RA, Boyce AJ, Clegg JB . Direct measurement of the male recombination fraction in the human beta-globin hot spot. Hum Mol Genet 2002; 11: 207–215.
Wainscoat JS, Hill AV, Boyce AL, Flint J, Hernandez M, Thein SL, Old JM, Lynch JR, Falusi AG, Weatherall DJ, Clegg JB . Evolutionary relationships of human populations from an analysis of nuclear DNA polymorphisms. Nature 1986; 319: 491–493.
Chen LZ, Easteal S, Board PG, Kirk RL . Evolution of beta-globin haplotypes in human populations. Mol Biol Evol 1990; 7: 423–437.
Long JC, Chakravarti A, Boehm CD, Antonarakis S, Kazazian HH . Phylogeny of human beta-globin haplotypes its implications for recent human evolution. Am J Phys Anthropol 1990; 81: 113–130.
Fullerton SM, Clegg JB . Hpa I,Hind III,Bam HI polymorphisms 3′ of the human beta-globin gene can be detected by a single polymerase chain reaction amplification product. Am J Hematol 1994; 47: 256.
Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB . Archaic African Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet 1997; 60: 772–789.
Harding RM, Fullerton SM, Griffiths RC, Clegg JB . A gene tree for beta-globin sequences from Melanesia. J Mol Evol 1997; 44( suppl 1): S133–S138.
Serjeant GR . Sickle-cell disease. Lancet 1997; 350: 725–730.
Chui DH, Dover GJ . Sickle cell disease: no longer a single gene disorder. Curr Opin Pediatr 2001; 13: 22–27.
Nagel RL, Fabry ME, Pagnier J, Zohoun I, Wajcman H, Baudin V, Labie D . Hematologically genetically distinct forms of sickle cell anemia in Africa: the Senegal type the Benin type. N Engl J Med 1985; 312: 880–884.
Nagel RL, Rao SK, Dunda-Belkhodja O, Connolly MM, Fabry ME, Georges A, Krishnamoorthy R, Labie D . The hematologic characteristics of sickle cell anemia bearing the Bantu haplotype: the relationship between G gamma HbF level. Blood 1987; 69: 1026–1030.
Nagel RL, Erlingsson S, Fabry ME, Croizat H, Susuka SM, Lachman H, Sutton M, Driscoll C, Bouhassira E, Billet HH . The Senegal DAN haplotype is associated with the amelioration of anemia in African-American sickle cell anemia patients. Blood 1991; 77: 1371–1375.
Kan YW, Dozy AM . Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc Natl Acad Sci U S A 1978; 75: 5631–5635.
Feldenzer J, Mears JG, Burns AL, Natta C, Bank A . Heterogeneity of DNA fragments associated with the sickle-globin gene. J Clin Invest 1979; 64: 751–755.
Panny SR, Scott AF, Smith KD, Phillips JA, Kazazian HH, Talbot CC, Boehm CD . Population heterogeneity of the Hpa I restriction site associated with the beta globin gene: implications for prenatal diagnosis. Am J Hum Genet 1981; 33: 25–35.
Hattori Y, Kutlar F, Kutlar A, McKie VC, Huisman TH . Haplotypes of beta S chromosomes among patients with sickle cell anemia from Georgia. Hemoglobin 1986; 10: 623–642.
Schroeder WA, Powars DR, Kay LM, Chan LS, Huynh V, Shelton JB, Shelton JR . Beta-cluster haplotypes, alpha-gene status, hematological data from SS, SC, S-beta-thalassemia patients in southern California. Hemoglobin 1989; 13: 325–353.
Oner C, Dimovski AJ, Olivieri NF, Schiliro G, Codrington JF, Fattoum S, Adekile AD, Oner R, Yuregir GT, Altay C, Gurgey A, Gupta RB, Jogessar VB, Kitundu MN, Loukopoulos D, Tamagnini GP, Ribeiro ML, Kutlar F, Gu LH, Lanclos KD, Huisman TH . Beta-S haplotypes in various world populations. Hum Genet 1992; 89: 99–104.
Centers for Disease Control and Prevention. Update: newborn screening for sickle cell disease—California, Illinois, New York, 1998. MMWR Morb Mortal Wkly Rep 2000; 49: 729–731.
Ventura SJ, Martin JA, Mathews TJ, Clarke SC . Advance report of final natality statistics, 1994. Monthly Vital Statistics Report, Vol 44, No 11, suppl. Hyattsville, MD: National Center for Health Statistics, 1996: 1–87.
Caggana M, Conroy JM, Pass KA . Rapid, efficient method for multiplex amplification from filter paper. Hum Mutat 1998; 11: 404–409.
Statistical Analysis Battery for Epidemiologic Research (SABER), Version 1.96 [computer program]. Atlanta, GA: Centers for Disease Control and Prevention, 1996.
Flint J, Harding RM, Boyce AJ, Clegg JB . The population genetics of the haemoglobinopathies. Baillieres Best Pract Res Clin Haematol 1998; 11: 1–51.
Arends A, Alvarez M, Velazquez D, Bravo M, Salazar R, Guevara JM, Castillo O . Determination of beta-globin gene cluster haplotypes prevalence of alpha-thalassemia in sickle cell anemia patients in Venezuela. Am J Hematol 2000; 64: 87–90.
Muniz A, Corral L, Alaez C, Svarch E, Espinosa E, Carbonell N, di Leo R, Felicetti L, Nagel RL, Martinez G . Sickle cell anemia beta-gene cluster haplotypes in Cuba. Am J Hematol 1995; 49: 163–164.
Keclard L, Romana M, Lavocat E, Saint-Martin C, Berchel C, Merault G . Sickle cell disorder, beta-globin gene cluster haplotypes alpha-thalassemia in neonates adults from Guadeloupe. Am J Hematol 1997; 55: 24–27.
Cuellar-Ambrosi F, Mondragon MC, Figueroa M, Prehu C, Galacteros F, Ruiz-Linares A . Sickle cell anemia beta-globin gene cluster haplotypes in Colombia. Hemoglobin 2000; 24: 221–225.
Goncalves I, Goncalves J, Perichon B, Osorio-Almeida L, Krishnamoorthy R, Lavinha J . A novel mosaic Bantu/Benin/Bantu beta s haplotype found in several African populations. Hum Genet 1994; 94: 101–103.
Kidd KK, Morar B, Castiglione CM, Zhao H, Pakstis AJ, Speed WC, Bonne-Tamir B, Lu RB, Goldman D, Lee C, Nam YS, Grandy DK, Jenkins T, Kidd JR . A global survey of haplotype frequencies linkage disequilibrium at the DRD2 locus. Hum Genet 1998; 103: 11–27.
Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, Kidd JR, Pakstis AJ, Jenkins T, Kidd KK . A global haplotype analysis of the myotonic dystrophy locus: implications for the evolution of modern humans for the origin of myotonic dystrophy mutations. Am J Hum Genet 1998; 62: 1389–1402.
Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, Batzer MA . The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, Y-chromosome data. Am J Hum Genet 2000; 66: 979–988.
Kidd JR, Pakstis AJ, Zhao H, Lu R, Okonofua E, Odunsi A, Grigorenko E, Bonne-Tamir B, Friedlaender J, Schulz LO, Parnas J, Kidd KK . Haplotypes linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 2000; 66: 1882–1899.
Peterson RJ, Goldman D, Long JC . Effects of worldwide population subdivision on ALDH2 linkage disequilibrium. Genome Res 1999; 9: 844–852.
Schmidley AD, Gibson C . Profile of the foreign-born population in the United States. Current Population Reports 1999. Washington, DC: US Government Printing Office. US Census Bureau, Series P23–195, 1–73.
Hanis CL, Hewett-Emmett D, Bertin TK, Schull WJ . Origins of US Hispanics: implications for diabetes. Diabetes Care 1991; 14: 618–627.
Therrien M, Ramirez RR . The Hispanic population in the United States: March 2000. Current Population Reports 2000. Washington, DC: US Government Printing Office. US Census Bureau, Series P20–535, 1–8.
Hiti AL, Zeng L, Xiang Q, Lorey FW, Powars DR . Beta-globin haplotypes from blood spots for follow-up of newborn hemoglobinopathy screening. Am J Hematol 1997; 54: 76–78.
Douglas JA, Boehnke M, Gillanders E, Trent JM, Gruber SB . Experimentally-derived haplotypes substantially increase the efficiency of linkage disequilibrium studies. Nat Genet 2001; 28: 361–364.
Ruano G, Kidd KK . Direct haplotyping of chromosomal segments from multiple heterozygotes via allele-specific PCR amplification. Nucleic Acids Res 1989; 17: 8392.
Therrell BL, Hannon WH, Pass KA, Lorey F, Brokopp C, Eckman J, Glass M, Heidenreich R, Kinney S, Kling S, Landenburger G, Meaney FJ, McCabe ERB, Panny S, Schwartz M, Shapira E . Guidelines for the retention, storage, use of residual dried blood spot samples after newborn screening analysis: statement of the Council of Regional Networks for Genetic Services. Biochem Mol Med 1996; 57: 116–124.
Andresen BS, Dobrowolski SF, O’Reilly L, Muenzer J, McCandless SE, Frazier DM, Udvari S, Bross P, Knudsen I, Banas R, Chace DH, Engel P, Naylor EW, Gregersen N . Medium-chain acyl-CoA dehydrogenase (MCAD) mutations identified by MS/MS-based prospective screening of newborns differs from those observed in patients with clinical symptoms: identification characterization of a new, prevalent mutation that results in mild MCAD deficiency. Am J Hum Genet 2001; 68: 1408–1418.
Pourfarzam M, Morris A, Appleton M, Craft A, Bartlett K . Neonatal screening for medium-chain acyl-CoA dehydrogenase deficiency. Lancet 2001; 358: 1063–1064.
Caggana M, Walker K, Reilly AA, Conroy JM, Duva S, Walsh AC . Population-based studies reveal differences in the allelic frequencies of two functionally significant human interleukin-4 receptor polymorphisms in several ethnic groups. Genet Med 1999; 1: 267–271.
Conroy JM, Trivedi G, Sovd T, Caggana M . The allele frequency of mutations in four genes that confer enhanced susceptibility to venous thromboembolism in an unselected group of New York State newborns. Thromb Res 2000; 99: 317–324.
Olivarez L, Caggana M, Pass KA, Ferguson P, Brewer GJ . Estimate of the frequency of Wilson’s disease in the US Caucasian population: a mutation analysis approach. Ann Hum Genet 2001; 65: 459–463.
Acknowledgements
The authors thank Dr. Muin Khoury and Sydney Kling for their work in defining the collaborative project and collecting the data. They also thank Salvatore Duva and Dr. James Conroy for their expert technical assistance. Oligonucleotides were synthesized at the Molecular Genetics Core Facility at the Wadsworth Center.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Crawford, D., Caggana, M., Harris, K. et al. Characterization of β-globin haplotypes using blood spots from a population-based cohort of newborns with homozygous HbS. Genet Med 4, 328–335 (2002). https://doi.org/10.1097/00125817-200209000-00003
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/00125817-200209000-00003