Abstract
Purpose
To study the molecular pathogenesis of a Chinese family with coronary form of cataract.
Methods
One Chinese three-generation family with inherited coronary cataract phenotype was recruited. Five affected and seven unaffected family members attended our study. Genome-wide linkage analysis was applied to map the disease loci, and two candidate genes from a locus on chromosome 1 and a locus on chromosome 22 were sequenced for mutation identification. Software at the Expasy proteomics server was utilized to predict the mutation effect on proteins.
Results
Whole genome linkage analysis indicated some regions on chromosome 1, 10, and 22, with LOD score values greater than 1. Within these loci, the GJA8 and CRYBB2 genes, located in the two loci with the highest LOD score of 1.51 on chromosomes 1 and 22, respectively, were sequenced. A novel mutation c.92C>G in exon 2 of CRYBB2 causing S31W was identified in all five patients. It was not found in 95 unrelated controls. This missense sequence alteration likely enhanced the local solubility. Around the mutation site, a lipocalin signature motif was predicted by ScanProsite.
Conclusions
A novel disease-causing mutation S31W in CRYBB2 was identified in a Chinese cataract family. It is the first reported mutation for coronary cataract. Functional characterization should be carried out to evaluate the biological effects of this mutant.
Similar content being viewed by others
Introduction
Congenital cataract refers to lens opacity presented at birth or shortly thereafter. It is responsible for approximately one-tenth of childhood blindness worldwide, with prevalence estimated to be about 1–6 per 10 000 live births in most populations.1, 2 Autosomal dominant congenital cataract (ADCC) is the major inherited mode of congenital cataract. So far, at least 18 genes with more than 60 mutations are known to be responsible for isolated ADCC that is not associated with systemic diseases.3 Functions and properties of these genes are widely diversified. They include nine crystallin genes: CRYAA, CRYAB, CRYBB1, CRYBB2, CRYBA1, CRYBA4, CRYGC, CRYGD CRYGS; four membrane transport protein genes: MIP, TMEM114,4 GJA8, GJA3; one cytoskeletal protein gene: BFSP2; three transcription factor genes: PITX3, MAF, and HSF4; and one chromatin modifying protein gene: CHMP4B.5
Crystallins account for 90% of water-soluble proteins in the human lens. After synthesis, they exist throughout the lifespan of the host without turnover in mature lens fiber cells. The correct expression of native crystallins and maintenance of their native properties against metabolic and environmental insult are crucial for lens transparency. There are three crystallin families: α-, β-, and γ-crystallins. α-Crystallins (CRYAA) that account for more than half of the total crystallins in human lens.6 Apart from being structural proteins that help to maintain light refraction through the lens, they also function as chaperones during lens development.7 Human β- and γ-crystallins belong to a super family and they share highly homologous sequences.
β-crystallins can be further classified into the acidic and basic subtypes. Acidic β-crystallins include βA1-, βA2-, βA3-, βA4-crystallins (CRYBA1, CRYBA2, CRYBA3, and CRYBA4, respectively), and basic β-crystallins include βB1-, βB2-, βB3-crystallins (CRYBB1, CRYBB2, and CRYBB3, respectively). CRYBB2 is the most abundant β-crystallin.6 It is also the most soluble and least modified crystalline protein during aging.8, 9 During the first year of life, the relative amounts of CRYBB2 and CRYAA in lens change drastically. CRYBB2 increases from 12 to 24% and αA-crystallin decreases from 30 to 18%, with a total percentage maintained at 42%. It seems that CRYBB2 is replacing CRYAA.8 CRYBB2 likely plays a contributive role in lens development, but the functional significance is not yet known.
In this study, we investigated the gene lesion that is associated with inherited coronary cataract in a Chinese family.
Materials and methods
Sample collection and DNA extraction
A Chinese family with autosomal dominant inherited cataract was recruited at the First Affiliated Hospital, Medical College, Zhejiang University, People's Republic of China. This study followed the tenets of Declaration of Helsinki. Twelve family members attended our study and signed consent forms. All of them were given detailed ophthalmic examinations, including visual acuity, intraocular pressure, slit lamp assessment, and indirect ophthalmoscopy under dilated pupils. Peripheral venous blood samples were collected, and genomic DNA was extracted using QIAamp DNA kit (Qiagen, Valencia, CA, USA). A total of 95 unrelated control subjects who were aged more than 50 years with no family history of congenital cataracts were also recruited. They did not have eye diseases except mild myopia and senile cataracts. They were given complete ophthalmologic examinations as the study subjects of the inherited cataract family.
Genome-wide linkage analysis
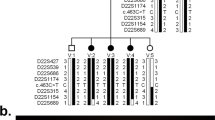
Whole genome scanning with 541 microsatellite markers having an average spacing of around 8 cM was carried out at deCODE (Sturlugata, Reykjavik, Iceland). Briefly, PCR samples were set up in multiplex with fluorescently labeled primers. When the PCR was completed, 12 markers were pooled for each individual sample. The amplified DNA fragments were analysed by capillary electrophoresis on an ABI 3730 DNA analyzer. The proprietary deCODE Allele Caller software was used for automated allele calling. The relationship between family members and genotyping error was examined by PedCheck, which identifies genotype incompatibilities in linkage analysis.10 Two-point linkage analysis was conducted by MLINK between disease and markers on chromosomes 1–22. MLINK subprogram was from FASTLINKAGE version 4.1P package. A gene frequency of 0.0001 and a penetrance of 100% were assumed for ADCC.
Gene sequencing
GJA8 and CRYBB2 were selected from the regions showing likely association with cataract as indicated by the whole genome scanning. All exons and adjacent splicing regions were sequenced with specific primers (Table 1) and BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) on an ABI PRISM™ 3130xl genetic analyzer (Applied Biosystems). The sequences were compared with references obtained from NCBI GeneBank (http://www.ncbi.nlm.nih.gov/Genebank/; NCBI accession numbers NM005267.3 and NM000496.2).
Computational methods
The effects of S31W on proteins were assessed using softwares at the Expasy proteomics server (http://ca.expasy.org/). Protein isoelectric point (pI) and molecular weight (MW) were calculated by the program of Compute pI/MW. The protein hydrophobicity was predicted by ProtScale. The polypeptide sequences were scanned for occurrence of specific motifs by ScanProsite at Prosite. The Protein 3D structure was obtained form RCSB protein data bank (http://www.rcsb.org/pdb/home/home.do) (PDB ID: 1ytq).
Results
Clinical phenotypes
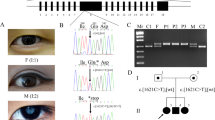
The inherited pattern of this family was consistent with autosomal dominant mode (Figure 1a). The age of diagnosis of disease ranged from 8 to 35 years. Five family members had bilateral inherited cataract and three of them had undergone lens extraction surgeries. The remaining two patients who did not undergo cataract surgery showed round opacity distributed in the deep cortex, which surrounded the nucleus like a crown (Figure 1b). The severity of cataract in both patients was similar. Their uncorrected visual acuities were 0.12–0.8 (Table 2). The diagnosis of bilateral coronary cataract was given by a senior ophthalmologist. None of the five affected family members have other ocular or systemic abnormalities. Other family members did not have lens opacity except for I2, who had senile cataract.
An inherited coronary cataract family. (a) The pedigree; (b) the lens picture taken from the patient of β1, a 24-year-old man, the age of diagnosis was 14 years. He showed round opacity distributed in the deep cortex, which surrounded the nucleus like a crown. The severity of cataract in both two lenses was similar, and the visual acuity of both eyes was 0.8.
Linkage analysis
Genomewide scanning mapped some promising regions on chromosome 1, 10, and 22 with a series of logarithm of the odds (LOD) scores bigger than 1.0. (Table 3). As the family size was not big, the highest LOD scores obtained on chromosomes 1 and 22 were only 1.51. Two possible loci were noted, one on chromosome 1 flanked by markers D1S484 and D1S452, the other on chromosome 22 flanked by D22S427 and D22S539. Two candidate genes reported to be responsible for ADCC, GJA8 and CRYBB2, were selected from these two regions for sequence analysis.
Mutation identification
Comparing with the CRYBB2 reference sequence (NCBI accession number NM000496.2), a new transversion from C to G was identified at the 92nd position (c.92C>G) in exon 2. It changed the 31st residue from serine to tryptophan (S31W) (Figure 2). The heterozygous substitution of S31W was observed in all patients, but not in 95 unrelated controls. Except for a 3-year-old unaffected girl (III3), other unaffected family members did not carry this mutation. No sequence variation was identified in the GJA8 gene.
Prediction of mutation effect on protein properties
The isoelectric point (pI) of both wild-type and mutant S31W was 6.5. The molecular weight of the mutant protein is about 23 479 Da, which was slightly higher than that of wild-type 23 380 Da. The hydrophobicity at the mutant site and the neighbouring regions obviously increased (Figure 3). Through ScanProsite, a new lipocalin signature motif was predicted in S31W mutant protein but not in wild type (Table 4). The serine at the position of 31 is located on the surface of N-terminal domain.
Discussion
We identified a novel disease causing mutation S31W in CRYBB2 in a Chinese cataract family. This missense variation was found in all the affected family members. Neither unrelated controls nor phenotypically normal family members carried this sequence change, except for a young girl who was 3 years old. Although this may suggest incomplete penetrance of disease, we noticed that the diagnosis age of patients in this family ranged from 8 to 35 years. It is still possible that the girl carrying the S31W mutation may later develop cataract. Thus, her lens transparency will be carefully followed in the future. Some ADCC genes, such as CRYAA, CRYGD, BFSP2, and MAF, have been reported to cause juvenile onset cataract. G98R in CRYAA was associated with juvenile onset cataract advancing from a peripheral ring-like opacity to a total cataract.11 R14C in CRYGD was responsible for a progressive juvenile onset punctuate cataract.12 E233del in BFSP2 was related to early childhood cataract and myopia.13 Another mutation in this gene R287W caused juvenile onset lamellar cataract.14 Sequence variations in MAF have also been shown to be involved in juvenile onset cataract.15 In one Indian family with autosomal recessive cataract, the deletion mutation of BFSP1 was reported to cause juvenile onset cataract.16 Another autosomal recessive cataract family showed an adult-onset lens opacity. The putative genetic lesion has been mapped in the region of 9q13-22.17
Including S31W, totally five mutations in CRYBB2 had been reported to be associated with various phenotypes of congenital cataract. In ADCC, W151C caused central nuclear opacity.18 Q155X led to diverse phenotypes, including cerulean, Coppock-like, polymorphic, and sutural cataracts.19, 20, 21, 22 D128V resulted in bilateral nuclear cataract surrounded by cortical opacity23 A homozygous 168G deletion, also in exon 2, of CRYBB2 caused a frameshift leading to a missense mutation at amino acid 57 and a truncated protein due to a stop codon at 107. It was responsible for bilateral confluent nuclear congenital cataract in an autosomal recessive mode of inheritance in offspring of consanguinity in two unrelated Israeli Bedouin families, indicating possible founder effect.24 The morphology of coronary cataract is distinctive from all these various cataract forms. Thus, our patients provided a new phenotype related to CRYBB2. Moreover, S31W is the first mutation reported for coronary cataract, which is characterized by club-shaped or oval opacities distributed in a radial pattern surrounding the nucleus like a crown. We have no evidence of founder mutation.
CRYBB2 is a soluble structural protein in human lens. It self-aggregates into homo-dimers or associates with other β-crystallins to form hetro-oligomers.21 Each CRYBB2 subunit, similar to monomeric CRYGD, has two tightly folded domains, N-terminal and C-terminal domains, composed of two Greek key motifs. However, the two domains are separated by an extended connecting peptide between the N-terminal domain of one subunit and the C-terminal domain of the other.25 Thus there are a lot of intermolecular contacts between domains in CRYBB2 compared with intramolecular contacts of CRYGD. Any mutation affecting this intermolecular action will lead to the change of solubility and stability of CRYBB2. W151C has been predicted to destroy the fourth Greek key motif and increase the protein hydrophobicity, which might affect the solubility of the mutant CRYBB2.17 Another mutation, Q155X, showed partial unfolded structure and decreased structure order, with reduced interactions with other proteins.26 D128V mutant was supposed to cause the random coil region between amino acids 126–139 of the mutant protein to become hydrophobic and electropositive.23
S31W did not affect the protein pI but greatly increased the local hydrophobicity around the mutant site. Three-dimensional analysis based on the 1ytq model showed that serine at position 31 was on the surface of N-terminal domain. It is possible that the substitution at this location alters the hydrophobic interactions among CRYBB2 molecules. Moreover, it creates a lipocalin signature motif in the mutant protein. The lipocalins share characteristic conserved sequence motifs and bind small hydrophobic molecules, such as steroids, bilins, retinoids, and lipids.27 They are also capable to bind cell-surface receptors and form complexes with soluble macromolecules.28 Lens clarity is a result of regular packing of water soluble proteins. CRYBB2 as a structural protein plays a key role in maintaining lens transparency. We thus hypothesize that the novel mutation S31W enhanced hydrophobicity and created a calipolin signature motif that could alter the local binding ability, which would disrupt dimerization of the CRYBB2 protein or impair binding with other lens-soluble proteins. This alteration may destroy the microstructure of lens and increase light scattering, leading ultimately to lens opacity.
In summary, a novel cataract-causing mutation, c.92C>G in exon 2 of the CRYBB2 gene, which causes the 31st residue serine substituted by tryptophan (S31W), was identified in an autosomal dominant coronary cataract family. This variant likely causes cataract through obviously enhanced local hydrophobicity and formed a new lipocalin signature motif. This is the first reported mutation for coronary cataract phenotype.
Accession codes
References
Francis PJ, Berry V, Bhattacharya SS, Moore AT . The genetics of childhood cataract. J Med Genet 2000; 37: 481–488.
Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT . Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol 2004; 49: 300–315.
Shiels A, Hejtmancik JF . Genetic origins of cataract. Arch Ophthalmol 2007; 125: 165–173.
Jamieson RV, Farrar N, Stewart K, Perveen R, Mihelec M, Carette M et al. Characterization of a familial t(16;22) balanced translocation associated with congenital cataract leads to identification of a novel gene, TMEM114, expressed in the lens and disrupted by the translocation. Hum Mutat 2007; 28: 968–977.
Shiels A, Bennett TM, Knopf HL, Yamada K, Yoshiura K, Niikawa N et al. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet 2007; 81: 596–606.
Robinson NE, Lampi KJ, Speir JP, Kruppa G, Easterling M, Robinson AB . Quantitative measurement of young human eye lens crystallins by direct injection Fourier transform ion cyclotron resonance mass spectrometry. Mol Vis 2006; 12: 704–711.
Horwitz J . Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad USA 1992; 89: 10449–10453.
Feng J, Smith DL, Smith JB . Human lens beta-crystallin solubility. J Biol Chem 2000; 275: 11585–11590.
Zhang Z, David LL, Smith DL, Smith JB . Resistance of human betaB2-crystallin to in vivo modification. Exp Eye Res 2001; 73: 203–211.
O'Connell JR, Weeks DE . PedCheck. a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998; 63: 259–266.
Santhiya ST, Soker T, Klopp N, Illig T, Prakash MV, Selvaraj B et al. Identification of a novel, putative cataract-causing allele in CRYAA (G98R) in an Indian family. Mol Vis 2006; 12: 768–773.
Stephan DA, Gillanders E, Vanderveen D, Freas-Lutz D, Wistow G, Baxevanis AD et al. Progressive juvenile-onset punctate cataracts caused by mutation of the gammaD-crystallin gene. Proc Natl Acad Sci USA 1999; 96: 1008–1012.
Zhang Q, Guo X, Xiao X, Yi J, Jia X, Hejtmancik JF . Clinical description and genome wide linkage study of Y-sutural cataract and myopia in a Chinese family. Mol Vis 2004; 17: 890–900.
Conley YP, Erturk D, Keverline A, Mah TS, Keravala A, Barnes LR et al. A juvenile-onset, progressive cataract locus on chromosome 3q21-q22 is associated with a missense mutation in the beaded filament structural protein-2. Am J Hum Genet 2000; 66: 1426–1431.
Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E et al. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet 2002; 11: 33–42.
Ramachandran RD, Perumalsamy V, Hejtmancik JF . Autosomal recessive juvenile onset cataract associated with mutation in BFSP1. Hum Genet 2007; 121: 475–482.
Heon E, Paterson AD, Fraser M, Billingsley G, Priston M, Balmer A et al. A progressive autosomal recessive cataract locus maps to chromosome 9q13-q22. Am J Hum Genet 2001; 68: 772–777.
Santhiya ST, Manisastry SM, Rawlley D, Malathi R, Anishetty S, Gopinath PM et al. Mutation analysis of congenital cataracts in Indian families: identification of SNPS and a new causative allele in CRYBB2 gene. Invest Ophthalmol Vis Sci 2004; 45: 3599–3607.
Gill D, Klose R, Munier FL, McFadden M, Priston M, Billingsley G et al. Genetic heterogeneity of the Coppock-like cataract: a mutation in CRYBB2 on chromosome 22q11.2. Invest Ophthalmol Vis Sci 2000; 41: 159–165.
Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P et al. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet 1997; 6: 665–668.
Vanita V, Sarhadi V, Reis A, Jung M, Singh D, Sperling K et al. A unique form of autosomal dominant cataract explained by gene conversion between beta-crystallin B2 and its pseudogene. J Med Genet 2001; 38: 392–396.
Yao K, Tang X, Shentu X, Wang K, Rao H, Xia K . Progressive polymorphic congenital cataract caused by a CRYBB2 mutation in a Chinese family. Mol Vis 2005; 11: 758–763.
Pauli S, Söker T, Klopp N, Illig T, Engel W, Graw J . Mutation analysis in a German family identified a new cataract-causing allele in the CRYBB2 gene. Mol Vis 2007; 13: 962–967.
Cohen D, Bar-Yosef U, Levy J, Gradstein L, Belfair N, Ofir R et al. Homozygous CRYBB1 deletion mutation underlies autosomal recessive congenital cataract. Invest Ophthalmol Vis Sci 2007; 48: 2208–2213.
Bax B, Lapatto R, Nalini V, Driessen H, Lindley PF, Mahadevan D et al. X-ray analysis of beta B2-crystallin and evolution of oligomeric lens proteins. Nature 1990; 347: 776–780.
Liu BF, Liang JJ . Interaction and biophysical properties of human lens Q155* betaB2-crystallin mutant. Mol Vis 2005; 11: 321–327.
Flower DR, North AC, Attwood TK . Structure and sequence relationships in the lipocalins and related proteins. Protein Sci 1993; 2: 753–761.
Flower DR . The lipocalin protein family. Structure and function. Biochem J 1996; 318: 1–14.
Acknowledgements
We acknowledge financial support by Scientific Research Grant of the Science and Technology Bureau 20070733B06, Hangzhou, Zhejiang Province.
Author information
Authors and Affiliations
Corresponding author
Additional information
Financial interest: None
Meeting presentation: None
Rights and permissions
About this article
Cite this article
Lou, D., Tong, JP., Zhang, LY. et al. A novel mutation in CRYBB2 responsible for inherited coronary cataract. Eye 23, 1213–1220 (2009). https://doi.org/10.1038/eye.2008.222
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/eye.2008.222