Abstract
Coenzyme Q10 (CoQ10) deficiency is associated to a variety of clinical phenotypes including neuromuscular and nephrotic disorders. We report two unrelated boys presenting encephalopathy, ataxia, and lactic acidosis, who died with necrotic lesions in different areas of brain. Levels of CoQ10 and complex II+III activity were increased in both skeletal muscle and fibroblasts, but it was a consequence of higher mitochondria mass measured as citrate synthase. In fibroblasts, oxygen consumption was also increased, whereas steady state ATP levels were decreased. Antioxidant enzymes such as NQO1 and MnSOD and mitochondrial marker VDAC were overexpressed. Mitochondria recycling markers Fis1 and mitofusin, and mtDNA regulatory Tfam were reduced. Exome sequencing showed mutations in PDHA1 in the first patient and in PDHB in the second. These genes encode subunits of pyruvate dehydrogenase complex (PDH) that could explain the compensatory increase of CoQ10 and a defect of mitochondrial homeostasis. These two cases describe, for the first time, a mitochondrial disease caused by PDH defects associated with unbalanced of both CoQ10 content and mitochondria homeostasis, which severely affects the brain. Both CoQ10 and mitochondria homeostasis appears as new markers for PDH associated mitochondrial disorders.
Similar content being viewed by others
Introduction
CoQ10 is a redox lipophilic molecule mainly located in inner mitochondria membrane that transfers electrons from complex I, and complex II to complex III in respiratory chain and participates in the superassembly of these complexes.1 It has antioxidant properties, regulates both transition pore and uncoupling proteins, and it is an obligated substrate for both β-oxidation of fatty acids and pyrimidine nucleotide biosynthesis.2 CoQ10 deficiency impairs oxidative phosphorylation and causes clinically heterogeneous mitochondrial diseases,3 including an encephalomyopathy form with muscle disorders and myoglobinuria; an ataxic form with atrophic cerebellum; a severe infantile form with renal failure; an isolated myopathy, and nephrotic syndrome, have been so far described.4 Primary deficiencies are due to mutations of genes involved in its biosynthesis, a very complex pathway not completely known, whose regulation has started to be described.5 In mammals, CoQ levels are tissue specific and its concentration in mitochondria is very much balanced with electron chain complexes, contributing to prevent endogenous oxidative stress.6 Among tissues, CoQ content in brain is very stable and highly independent of environment.7 Furthermore, mutations of genes that are not part of respiratory chain also cause mitochondrial dysfunctions and may induce an adaptive decrease of CoQ10, named secondary deficiency, although the mechanisms are totally unknown.4, 5
In contrast to the already described cases of mitochondrial diseases showing CoQ10 deficiency, we describe here two unrelated children that present a fatal neurological disorder, associated to genetic variants in PDH genes. The two cases presented high levels of both CoQ10 and mitochondria in fibroblasts and skeletal muscle, with low efficient mitochondria that show defective homeostasis, and high levels of antioxidant enzymes.
Subjects and methods
Patient 1
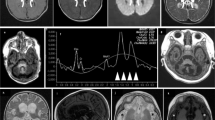
This was a son from nonconsanguineous parents, with a negative family history. The parents had had two previous miscarriages. The boy presented the first clinical manifestations at 10 months old, showing hypertonus that lasted 1 week, with no other symptoms. One month later, during a viral process, he presented drowsiness, hypotonia, bilateral ptosis and ataxia, with a complete recovery after 15 days. At 18 months of age, after a febrile episode, he manifested ataxia, hypertonus and limitations of the upgaze; blood lactate values were markedly elevated. MRI evidenced basal ganglia lesions, in cerebellar pedunculi and bulbomedullar junction. In this occasion, after the resolution of the acute process, the patient manifested decreased strength in the right part of the body. Under the suspicion of a mitochondrial disorder, muscle biopsy was performed. At age 2 years, he presented a new episode of acute ataxia with increased lactate values in both, blood and cerebrospinal fluid after which chronic ataxia persisted. Subsequently he presented repeated episodes, characterized by vomiting, worsening of ataxia, dysarthria, alteration of cranial nerves, loss of consciousness and hyperlactacidemia triggered by stress, physical exercise or febrile episodes. There was a progressive worsening of the general clinical condition during intercritical periods. Electromyography showed axonal neuropathy. The patient died at age 9 years and 5 months, after an acute febrile process, which triggered tonic–clonic seizures, loss of consciousness and marked brainstem dysfunction. MRI evidenced necrotic lesions in cortical, subcortical and supratentorial areas at basal ganglia, cerebellum, bulb, colliculi and mesencephalus.
Patient 2
He was the first son of nonconsanguineous parents, with a negative family history. Pregnancy and perinatality were normal. The first clinical signs appeared at age 16 months with hypotonia, gait instability and vomiting during an upper respiratory tract infection, which resolved spontaneously without reliquates. At age 2 years and 6 months he was admitted to the hospital because of acute ataxia triggered by another upper respiratory tract infection. During infancy, he presented several other similar episodes during intercurrent diseases, characterized by acute ataxia, dysarthria, ptosis and acufens lasting from hours to several days. In the course of such episodes, both lactate and pyruvate values were increased in both blood and cerebrospinal fluid. Muscle biopsy was performed at age 3 years. Mild ataxia and mental disability became evident at age 11. Brain MRI showed T2 bilateral hyperintensities in lenticular nuclei, bulbar and right cerebellar hemisphere. Electroretinogram was normal. At age 16 years, during an infectious process, which evolved to sepsis, the patient developed severe lactic acidosis and brainstem dysfunction (with necrotic lesions in MRI), which rapidly lead to death.
Standard protocol approvals, registrations and patient consents
The Ethical Committee of Hospital Sant Joan de Déu approved the study. Written informed consent was obtained from the parents to conduct the study.
Muscle and skin biopsies
Muscle biopsies were collected and stored, and skin biopsies were cultured following for both the standardized procedures previously reported.8 No CoQ10 supplementation therapy was done prior to muscle or skin biopsies collection. All muscle biopsies were studied by standard histopathological procedures.
Fibroblasts culture
Control aged-matched human dermal fibroblasts (HDF) and patients skin fibroblasts (2 to 4 passes old) were cultured as described.9 Fibroblasts growth was determined in single plates for 144 h and counted each 24 h using a Neubaer chamber with an inverted microscope.
Mitochondrial biochemistry
CoQ10 levels were determined and CoQ10 biosynthesis rate was measured as the incorporation of the radiolabelled p-hydroxybenzoic acid ([14C]-pHB) as described.9 Activities of NADH:coenzyme Q1 oxidoreductase (complex I), cytochrome C oxidoreductase (complex III), NADH:cytochrome C reductase (complex I+III), succinate:cytochrome C reductase (complex II+ III) and citrate synthase (CS) were determined in cell extracts using previously described spectrophotometric methods.10 Results are expressed as specific activity (μmol/min/mg protein; mean±SD). Mitochondrial membrane potential (ΔΨm) and reactive oxygen species (ROS) generation were carried out as described.9 Proteins of fibroblasts homogenates were analyzed by the Lowry procedure.11
ATP determination
ATP level was determined by bioluminescence method using commercial Molecular Probes’ ATP Determination Kit (A22066). Assay is based on recombinant firefly luciferase requirement (and its substrate D-luciferin) for ATP in producing light emission maximum ~560 nm at pH 7.8 from the reaction: luciferin+ATP+O2 → (Mg2+ luciferase)→ oxyluciferin+AMP+pyrophosphate+CO2+light. This assay is able to detect as little as 0.1 picomol of ATP. Standard curves were run with each experiment.
Molecular studies
Total DNA preparation was carried out following standard procedures. Exomes were tested against the Single Nucleotide Polymorphism (dbSNP) and the 1000 genome project databases (1000 g) and the ESP 5400 data release of the NHLBI GO Exome Sequencing Project (ESP5400). ‘Sorting tolerant from intolerant’ (sift) and ‘polymorphism phenotyping’ (polyphen) algorithms were used to predict the consequences of the variations. Candidate variants were confirmed by standard sequencing procedure of genomic and/or cDNA fragments. RNA was extracted with easy-BLUE Total RNA Extraction kit (iNtRON Biotechnology) according to manufacturer's instructions. One microgram of total RNA was used to obtain cDNA of PDH genes with the iScript cDNA Sintesis Kit (Bio-Rad) following manufacturer's instructions. Reference sequences NG_016781.1 and NM_000284.3 for PDH1 and NG_016860.1 and NM_000925.3 for PDHB were used in this study. Variants and phenotypes described herein are accessible in the gene variant database at http://databases.lovd.nl/shared/screenings/0000032781 (individual ID 00032713) and http://databases.lovd.nl/shared/screenings/0000032784 (individual ID 00032716) for PDHA1 and PDHB respectively. Exons in PDHB gene are numbered as in NG_016860.1. Primers 5′-0ATGAGGAAGATGCTCGCC-3′ and 5′-TTAACTGACTGACTTAAACTTGATCC-3′ were used to PCR amplify and validate variant c.832G>A in PDHA1 cDNA from patient 1. Primers 5′-CCTGGAGGGCATGTCTGTTA-3′ and 5′-CAGGCTGGACAACAGAGTGA-3′ were used to confirm variant c.301A>G in PDHB gene from genomic DNA of patient 2. Primers 5′- GGTCACCGCCTACTGTCAAT-3′ and 5′-GCAGGGAGAGAGGAAGATGA-3′ were used to confirm variant c.42+1G>A in PDHB gene from genomic DNA of patient 2. To investigate the effect of the c.42+1G>A variant, a 521 bp cDNA fragment from PDHB gene in patient 2 was amplified with primers 5′-CTGGCTTGGTGCGGAGAC-3′ and 5′-CCAGGGACTGACCACCTTTA-3′ (primers F1 and R1 in Figure 2, respectively). Bands were purified and cloned in plasmid pYES 2.1 TOPO (Invitrogen, Madrid, Spain). Alternative splicing caused by c.42+1G>A variant was confirmed by PCR amplification of cloned fragments with primers F1, R1 and R2 (5′-GCAGGGAGAGAGGAAGATGA-3′) and later sequencing. See Figure 2e for a schematic representation of the strategy followed to prove alternative splicing of intron 1 due to variant c.42+1G>A in PDHB from patient 2. Mitochondrial DNA from both patients was sequenced by standard procedure.
Immunoblotting
Western blotting was performed using standard methods. After protein transfer, membrane was incubated with primary antibodies against NQO1 (sc-16464, Santa Cruz Biotechnology, Heidelberg, Germany), MnSOD (574596, Calbiochem, Barcelona, Spain), TFAM (sc-23588, Santa Cruz Biotechnology), PGC-1α (516557, Calbiochem), VDAC/Porin (AV35122, Sigma, Madrid, Spain), Mitofusin (ab57602, Abcam, Madrid, Spain) and Fis1 (ALX-210-1037-0100, Enzo Life Sciences, Inc., Madrid, Spain). Corresponding rabbit anti-goat (SC-2768, Santa Cruz Biotechnology), goat anti-rabbit (401353, Calbiochem), goat anti-mouse (401253, Calbiochem) or rabbit anti-sheep (402100, Calbiochem) secondary antibodies coupled to horseradish peroxidase were later applied. Specific protein complexes were identified using HRP substrate.
Statistical analysis
All results are expressed as mean±SD of three independent experiments. The measurements were statistically analyzed using Student t-test for comparing two groups. The level of significance was set at P≤0.05.
Results
Histology of muscle of P1 showed few muscle fibers with the presence of morphologically abnormal mitochondria with no other histopathological signs. Optic microscopy showed abnormal fibers with subsarcolemmal deposits suggestive of a mitochondrial disorder in P2 muscle. Muscle CoQ10 content was slightly increased in P1 (0.59 μmol/mg prot; controls: 0.11–0.48) and more significantly in P2 (0.84 μmol/mg of protein), which were also higher when referred to citrate synthase (Table 1). CoQ10 content and biosynthetic rates were significantly increased in fibroblasts of both patients that corresponded to the increase of mitochondrial mass (Table 1). Complex I+III activity was in the upper level of reference values in muscle of both patients, and was significantly higher in fibroblast from P1. Complex II+III activity was also at the upper reference value in P1 muscle and was significantly higher in both P2 muscle and fibroblasts of both patients (Table 1). Both activities corresponded to higher mitochondria in these fibroblasts as indicated when referred to citrate synthase. Complex IV activity was unaffected in patient’s fibroblasts (Table 1). These results correlated with a significant increase of both mitochondrial membrane potential and oxygen consumption in fibroblasts of both patients (Table 1). Lactate accumulation in media after 48 h growth was higher in P1 fibroblasts (82.9±4.8 a.u./cells number, P≤0.05) and P2 fibroblasts (75.6±7.0 a.u./cells number, P≤0.05) compared with control cells (45.2±6.6 a.u./cells number). The unbalanced mitochondria showed a low efficiency when ATP levels were referred to citrate synthase in these fibroblasts (Figure 1a), and patient’s fibroblasts also showed a higher growth rate than control (Figure 1b). The patients did not present deletions and depletions of the mtDNA and sequencing of the whole mtDNA did not show any pathological mutation. However, mtDNA function must be affected in both patients’ fibroblasts owing to the significant decrease of Tfam but not mitochondria biogenesis owing to the unchanged expression of PGC1α (Figure 1c). The increased level of VDAC/porine determined by immonoblotting in both fibroblasts confirmed the higher mitochondria mass determined by citrate synthase. Likewise, mitochondrial recycling was also affected as Fis1 and mitofusin were decreased in these fibroblasts (Figure 1c). In order to understand the physiology of these cells that are representative of somatic cells from patients, we analyzed reactive oxygen species (ROS) generation that did not show differences with control but antioxidant enzymes NQO1 and MnSOD were clearly increased explaining the low ROS concentration (Figure 1c).
Properties of skin fibroblasts of P1 and P2. (a). ATP content in control and patients fibroblasts. Results are expressed as mean±SD of three independent experiments. *P<0.05 between control and patients fibroblasts. (b). Fibroblasts proliferation at 144 h from seeding. Both control and patients’ fibroblasts were seeded at a initial concentration of 1,75*105 cells per well. Results are expressed as mean±SD of three independent experiments. *P<0.05 between control and patients fibroblasts. (c). Immunolocalization of antioxidant enzymes NADH quinone oxidoreductase (NQO1) and Manganese Superoxide dismutase (MnSOD); Mitochondrial protein VDAC/Porine; Mitochondrial transcription activator TFam and PGC1α; Pro-fission protein FIS1 and Pro-fusion protein Mitofusin. Ponceau staining reflects equal protein loading.
In order to approach for the molecular origin of the disease we developed the sequence of the whole exome of both patients (the complete sequence information is available upon request).
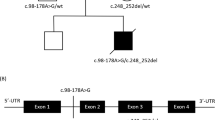
Variants were filtered following criteria of a minor allele frequency lower than 0,5% in the 1000 genome database and expected protein damage according to SIFT and Polyphen algorithms (see Subjects and Methods). In this way, variants compatible with the clinical symptoms of both patients were found in genes encoding for subunits of the pyruvate dehydrogenase complex. Patient 1 presented a c.832G>A substitution at the coding region of the PDHA1 gene, leading to a p(G278R) change at the E1α subunit of the PDH complex (Figure 2a). The p(G278R) change at PDHA1 has been previously described to affect the stability of the PDH complex resulting in a reduction by 60% of the total PDH activity in fibroblasts from patients.12 The c.832G>A variant in PDHA1 was a de novo mutation as it was absent in all, the parents and a healthy brother of patient 1. Patient 2 presented two variants in compound heterozygosis at the PDHB gene, which encodes the E1β subunit of the PDH complex. The c.301A>G variant, maternally inherited, led to a methionine to valine substitution at position 101 (Figure 2b). M101 is a highly conserved amino acid and p.M101V and p.M101T substitutions have been observed in other patients with reduced PDH activity in both, fibroblasts and skeletal muscle.13 The second variant, paternally inherited, is a novel c.42+1G>A substitution, affecting the first nucleotide of the first intron of the PDHB gene (Figure 2c). PCR amplification of a region comprising the first 500 bp of the PDHB cDNA resulted in double bands in patient 2. The detection of a weak upper band suggested an aberrant mRNA maturation compatible with the lack of splicing of the first intron and the consequent degradation of the messenger (Figure 2d). Both bands were cloned in pYES 2.1 TOPO plasmid and a second PCR was done using a forward primer aligned to the plasmid backbone and a reverse primer aligned to the end of first intron (Figure 2e). Amplification was only observed with the sample containing the cloned upper band (Figure 2f), supporting that the observed double band corresponded to alternatively spliced molecules of PDHB mRNA. The reverse primer used in this experiment aligned three nucleotides before the second exon (Figure 2e). This suggests that the c.42+1G>A variant does not promote the appearance of an alternative splicing site, but a complete absence of maturation of the first intron. This hypothesis was finally confirmed by direct sequencing of both cloned cDNA fragments, finding that the c.42+1G>A variant avoids the maturation of the first intron, resulting in a premature stop codon at the second exon (data not shown) and the consequent degradation of the affected mRNA molecules. Importantly, the c.42+1G>A variant was also found in a healthy brother of patient 2 whereas the variant c.301A>G was absent. This result, together with the single variants found in the parents of patient 2 confirms that PDHB deficiencies are associated to homozygous or compound heterozygous mutations.
Mutations in genes encoding for PDH complex subunits. (a). Sequencing of c.832G>A mutation (red square) in PDHA1 cDNA from patient 1. (b) Sequencing of maternally inherited c.301A>G mutation (red square) in PDHB cDNA, leading to p.M101V substitution. (c) Sequencing of paternally inherited substitution c.42+1G>A (red square) in genomic DNA, leading to altered splicing of intron of PDHB gene in patient 2. (d) PCR amplification of a 500 pb cDNA fragment of PDHB from patient 2 and control. Arrow indicates the alternatively maturated mRNA including the unspliced first intron. (e) Schematic representation of up and down fragments cloned in pYES 2.1 TOPO vector. Exons are represented as boxes, unspliced intron in UP fragment as a blue line and backbone of pYES 2.1 TOPO vector in orange. F1, R1 and R2 primers were used to prove the alternative splicing of PDHB gene in patient 2. (f) PCR amplification of Up and Down cloned cDNA fragments observed in patient 2 in figure (d). M, F and P2 in 2B and 2C panels correspond to mother, father and patient 2, respectively. The full colour version of this figure is available at European Journal of Human Genetics online.
Discussion
The content of CoQ10 in mitochondria is very much balanced with respiratory chain complexes contributing to their superassembly,1 which contributes to the efficiency of oxidative phosphorylation and limiting ROS production.6 Also, CoQ levels in brain are remarkably stable in mammals and are independent from environment.7 This is the base of understanding that a defective biosynthesis of CoQ10 by mutations in any COQ genes, and secondary deficiencies by defects of respiratory chain complexes functions such as mtDNA depletions14 or proteins involved in mitochondrial homeostasis cause mitochondrial diseases mainly with neuromuscular phenotype.5 The two patients presented here showed a similar phenotype to those described in encephalomyopathy, mainly ataxia, associated to primary or secondary CoQ10 deficiency but in these cases CoQ10 levels were elevated in both muscle and fibroblasts, corresponding to the increase of mitochondrial mass, showing mitochondrial dysfunction with low efficiency on ATP biosynthesis. A similar condition of defective respiration in the presence of high content of CoQ has been described in COQ10 null mutant yeast strain,15 indicating that CoQ is one major component in respiratory chain but requires proteins for its electron carrier transport to properly prevent defective electrons leaking for ROS production. Elevated levels of O2− have been observed in PDH deficient fibroblasts, probably produced at the Q0 site in complex III.16 We were unable to detect ROS production in fibroblasts of patients, however the overexpression of antioxidant enzymes MnSOD and NQO1, which is also a superoxide scavenger,17 demonstrates that electron leaking is indeed happening. Furthermore, NQO1 catalyzes NAD(P)H oxidation by reducing CoQ1018 that would compensate the redox balance of the high CoQ10 content. Also, the microscopy discovery of abnormal mitochondria correlates with the decrease of mitochondria recycling enzymes demonstrating the accumulation of low efficient mitochondria. The unbalanced mitochondria were also confirmed by the decrease of Tfam, which regulates expression and maintenance of mtDNA.19
The variants detected in PDH complex after exome sequencing would explain the clinical phenotype that has been previously described in patients with mutations in genes of this complex.13 In these cases, the rise of mitochondria and the corresponding CoQ10 concentration would try to compensate the lack of acetyl-CoA and also to contribute to NQO1 activity to prevent oxidative damage. These results indicate the importance of the balanced requirement for CoQ10 and electron chain complexes, and also promote that early diagnosis of CoQ10 content and citrate synthase are required to predict new clinical phenotypes of mitochondrial diseases.
References
Lapuente-Brun E, Moreno-Loshuertos R, Acín-Pérez R et al: Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 2013; 340: 1567–1570.
Bentinger M, Tekle M, Dallner G : Coenzyme Q—biosynthesis and functions. Biochem Biophys Res Commun 2010; 396: 74–79.
Emmanuele V, López LC, López L et al: Heterogeneity of coenzyme Q10 deficiency: patient study and literature review. Arch Neurol 2012; 69: 978–983.
Laredj LN, Licitra F, Puccio HM : The molecular genetics of coenzyme Q biosynthesis in health and disease. Biochimie 2014; 100: 78–87.
Doimo M, Desbats MA, Cerqua C, Cassina M, Trevisson E, Salviati L : Genetics of coenzyme Q10 deficiency. Mol Syndromol 2014; 5: 156–162.
Genova ML, Lenaz G : Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta 2014; 1837: 427–443.
Parrado-Fernández C, López-Lluch G, Rodríguez-Bies E et al: Calorie restriction modifies ubiquinone and COQ transcript levels in mouse tissues. Free Radic Biol Med 2011; 50: 1728–1736.
Montero R, Sánchez-Alcázar JA, Briones P et al: Analysis of coenzyme Q10 in muscle and fibroblasts for the diagnosis of CoQ10 deficiency syndromes. Clin Biochem 2008; 41: 697–700.
López-Martín JM, Salviati L, Trevisson E et al: Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum Mol Genet 2007; 16: 1091–1097.
Spinazzi M, Casarin A, Pertegato V, Salviati L, Angelini C : Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat Protoc 2012; 7: 1235–1246.
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ : Protein measurement with the Folin phenol reagent. J Biol Chem 1951; 193: 265–275.
Imbard A, Boutron A, Vequaud C et al: Molecular characterization of 82 patients with pyruvate dehydrogenase complex deficiency. Structural implications of novel amino acid substitutions in E1 protein. Mol Genet Metab 2011; 104: 507–516.
Quintana E, Mayr JA, García Silva MT et al: PDH E1β deficiency with novel mutations in two patients with Leigh syndrome. J Inherit Metab Dis 2009; 32 (Suppl 1): S339–S343.
Montero R, Grazina M, López-Gallardo E et al: Coenzyme Q10 deficiency in mitochondrial DNA depletion syndromes. Mitochondrion 2013; 13: 337–341.
Barros MH, Johnson A, Gin P, Marbois BN, Clarke CF, Tzagoloff A : The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J Biol Chem 2005; 280: 42627–42635.
Glushakova LG, Judge S, Cruz A, Pourang D, Mathews CE, Stacpoole PW : Increased superoxide accumulation in pyruvate dehydrogenase complex deficient fibroblasts. Mol Genet Metab 2011; 104: 255–260.
Siegel D, Gustafson DL, Dehn DL et al: NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol 2004; 65: 1238–1247.
Winski SL, Koutalos Y, Bentley DL, Ross D : Subcellular Localization of NAD(P)H:quinone Oxidoreductase 1 in Human Cancer Cells. Cancer Res 2002; 62: 1420–1424.
Gomes AP, Price NL, Ling AJY et al: Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 2013; 155: 1624–1638.
Acknowledgements
We thank the Bioinformatics Platform for Rare Diseases (CIBERER BIER), Valencia, Spain, for their help in the analysis of the exome sequencing data. This research was funded by grants PI14/00028 (RA), PI10-00662 (JM), and PI14/01962 (PN) from the Spanish Ministry of Health (Fondo de Investigación Sanitaria, Instituto de Salud Carlos III). LS was funded by Telethon Italy (GGP13222), the University of Padova (CPDA123573/12), and Fondazione CARIPARO (to LS). CA was funded by grant APP2D11086 from the University Pablo de Olavide (Plan Propio de Investigación).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Asencio, C., Rodríguez-Hernandez, M., Briones, P. et al. Severe encephalopathy associated to pyruvate dehydrogenase mutations and unbalanced coenzyme Q10 content. Eur J Hum Genet 24, 367–372 (2016). https://doi.org/10.1038/ejhg.2015.112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.112
This article is cited by
-
Cryo-EM snapshots of a native lysate provide structural insights into a metabolon-embedded transacetylase reaction
Nature Communications (2021)
-
CoQ10 a super-vitamin: review on application and biosynthesis
3 Biotech (2018)
-
Cerebrospinal fluid monoamines, pterins, and folate in patients with mitochondrial diseases: systematic review and hospital experience
Journal of Inherited Metabolic Disease (2018)