Abstract
We identified a novel homozygous 15q13.3 microdeletion in a young boy, with a complex neurodevelopmental disorder characterized by severe cerebral visual impairment with additional signs of congenital stationary night blindness, congenital hypotonia with areflexia, profound intellectual disability, and refractory epilepsy. The mechanisms by which the genes in the deleted region exert their effect are unclear. In this paper, we probed the role of downstream effects of the deletions as a contributing mechanism to the molecular basis of the observed phenotype. We analyzed gene expression of lymphoblastoid cells derived from peripheral blood of the proband and his relatives to ascertain the relative effects of the homozygous and heterozygous deletions. We identified 267 genes with apparent differential expression between the proband with the homozygous deletion and 3 age- and sex-matched typically developing controls. Several of the differentially expressed genes are known to influence neurodevelopment and muscular function, and thus may contribute to the observed cognitive impairment and hypotonia. We further investigated the role of CHRNA7 by measuring TNFα modulation (a potentially important pathway in regulating synaptic plasticity). We found that the cell line with the homozygous deletion lost the ability to inhibit the activation of tumor necrosis factor-α secretion. Our findings suggest downstream genes that may have been altered by the 15q13.3 homozygous deletion, and thus contributed to the severe developmental encephalopathy of the proband. Furthermore, we show that a potentially important pathway in learning and development is affected by the deletion of CHRNA7.
Similar content being viewed by others
Introduction
The 15q13.3 heterozygous microdeletion (OMIM number 612001) is a recognizable genomic spectrum disorder characterized by a range of neurodevelopmental disorders, including seizures, cognitive and social impairments, autism, and schizophrenia.1, 2, 3, 4, 5, 6, 7, 8 The phenotype varies in spectrum and severity owing to incomplete penetrance or variable expressivity. The 15q13.3 deletion is typically 1.6 Mb, harboring at least seven genes, ARHGAP11B, MTMR15, MTMR10, TRPM1, KLF13, OTUD7A, and CHRNA7. CHRNA7 codes for the α7 neuronal nicotinic receptor (α7nAChR).9 Alterations in α7nAChR are reported to cause seizures and neuropsychiatric disturbances in the affected individuals with an incomplete dominant inheritance pattern.4, 5, 6, 8, 10, 11, 12 One large genomic database survey identified 19 individuals with isolated heterozygous CHRNA7 gene deletions and 1 with an isolated homozygous deletion.13 The neurological symptoms in individuals with isolated CHRNA7 deletions implicate CHRNA7 as the major candidate gene responsible for the predominant manifestations of 15q13.3 microdeletion syndrome.
Six patients with homozygous deletions of 15q13.3 have been reported13, 14, 15, 16, 17 with a strikingly similar developmental encephalopathy that includes severe hypotonia, intractable epilepsy, and visual impairment. Although the CHRNA7 deletion is posited to cause seizures and neuropsychiatric disturbances, homozygous TRPM1 (transient receptor potential cation channel, subfamily M, member 1) deletion appears to cause visual impairment, in particular congenital stationary night blindness (CSNB).17, 18, 19, 20, 21
Although analysis of structural variants are beginning to provide insight into functional genomics,22 the exact relationships between altered CHRNA7 and TRPM1 gene expression, the associated abnormal phenotypes, and the roles of the other genes within the deleted region remain unclear. It is entirely possible that the pathogenic mechanisms underlying CHRNA7 and TRPM1 loss are a direct consequence of the deletion. However, there may be important contributions to the observed phenotype from downstream genes. We previously described a patient with a 15q13.3 homozygous microdeletion.15 This boy has a complex neurodevelopmental disorder, including severe cognitive impairment, congenital hypotonia with areflexia, and refractory epilepsy. The father, the mother, and the paternal grandmother were all heterozygous carriers of the deletion, and apparently clinically unaffected. This presented us with the unique opportunity to try to understand the molecular mechanisms that may contribute to the phenotype of patients with such deletions. We first asked whether the deletion resulted in a measurable, altered gene expression level. To study the effect of 15q13.3 deletion on gene expression, we compared gene expression profiles from lymphoblastoid cell lines from the proband with 15q13.3 homozygous deletion, heterozygous deletion family members, and age- and sex-matched controls, using a combination of whole-genome microarrays and bioinformatic tools. We then tested a possible monogenetic mechanism by testing whether CHRNA7 deletion causes loss of tumor necrosis factor-α (TNFα) modulation. TNFα is thought to have an important role in synaptic scaling.23 An inability to modulate TNFα may therefore account for the developmental impairments observed in patients with CHRNA7 deletion.
Materials and methods
Subjects
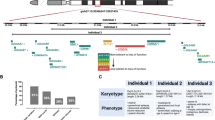
The proband with 15q13.3 homozygous deletion and his parents with 15q13.3 heterozygous deletions described in our previous report15 were evaluated using the Affymetrix aCGH-244K arrays (build hg18; Affymetrix Inc, Santa Clara, CA, USA). The aCGH data were validated by qPCR using primers specific to CHRNA7, as previously described.15 The extended family and comparison subjects were also screened using the CHRNA7 primers. Immortalized lymphoblastoid lines were created from the proband, his mother, father, a paternal half sister, a paternal half brother, a paternal grandmother, and two paternal great uncles. Three lymphoblastoid lines from age- and sex-matched normally developing comparison subjects with normal chromosomes were obtained from the Coriell cell repository ((AG14724 (8 years old), AG14948 (10 years old), AG14980 (9 years old)), and were used for gene expression comparison with the proband. The samples from the father, mother, and paternal grandmother with heterozygous 15q13.3 deletions were compared with samples from lymphoblastoid cells from adult male and female subjects, respectively (normal copy number for 15q13.3 determined by qPCR). The subject demographics are summarized in Table 1. In addition, we extracted RNA from an adult female brain (39 years old), to compare CHRNA7 expression level between lymphoblastoid cells and brain tissue.
Microarray data analysis
We used Affymetrix whole-genome exon microarrays (HuEx-1_0-st-v2, Affymetrix Inc) for gene expression analysis as previously described.24, 25 All quality assessments and signal intensities were adequate, and microarray data have been deposited in the Gene Expression Omnibus (NIH), accession number is GSE43583. Raw data (CEL files) were uploaded into the Partek Genomics Suite software version 6.5 (Partek Inc, St Louis, MO, USA) for normalization and statistical analysis. The Robust Multichip Analysis was used for background correction, followed by quantile normalization with baseline transformation to the median of the control samples. Probes with intensity values, above 20% of the background value in at least one of the conditions, were included for additional analysis. To overcome the limitation of having a single 15q13.3 homozygous deletion sample, we used a one-way ANOVA model using the Method of Moments to test for statistical significance.26 Genes with P-values≤0.01 were kept for further analysis.
The Ingenuity Pathways Analysis (IPA) version 9.0 (Ingenuity Systems, Inc, Redwood City, CA, USA) was used to explore associations between the differentially expressed gene data set and networks, canonical pathways, and predefined functional categories. IPA contains hundreds of pathways and identifies significant associations between the experimental data set and canonical pathways, functional categories, or disease associations within the database, by comparing the ratio of the number of molecules from the data set that map to a given pathway divided by the total number of molecules that map to the pathway. A Fisher’s exact test was used to calculate a P-value determining the probability that the association between the genes in the data set and the pathway was explained by chance alone. All biological functions and/or diseases in the IPA’s database were considered for the analysis without bias. Significance was defined as a P-value≤0.05.
Quantitative RT–PCR
Quantitative reverse transcription-PCR (QRT–PCR) was used to evaluate the expression of CHRNA7 and TRPM1, using gene-specific Taqman assays (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s directions. Lymphoblastoid cells were grown in RPMI 1640 media (Sigma, St Louis, MO, USA) supplemented with 10% fetal calf serum and 1% penn/strep. RNA was isolated from rapidly growing cell cultures using a mirVana RNA isolation kit (Invitrogen, Grand Island, NY, USA). RNA was reverse transcribed using 1 μg of total RNA from each sample using an ABI Superscript cDNA kit (Applied Biosystems). The cDNA was then PCR-amplified using human TaqMan probes (Applied Biosystems) for each sample. QRT–PCR was performed in triplicate on an ABI 7000 sequence detection instrument (Applied Biosystems). The point at which the intensity level crossed the PCR cycle threshold (CT) was used to compare individual reactions. Data were normalized to GAPDH, and results were shown as fold change relative to control subjects. All assays were run with negative control reactions with no template. All negative controls produced no detectable signal at 50 cycles.
Assessment of α7nAChR influence on TNFα response to lipopolysaccharide
We used the lymphoblastoid cells from our proband and two typically developing comparison males to assess whether activation of α7nAChR protein by nicotine could suppress the induction of TNFα expression in these cells as has been demonstrated previously in macrophages.27 Actively growing cells were treated with three different conditions as follows: Condition 1, 100 ng/ml lipopolysaccharide (LPS) for 4 h; Condition 2, 10 μM nicotine was added 10 min before the addition of 100 ng/ml LPS, followed by 4 h incubation; and Condition 3, 1 nM α-bungarotoxin (α-btx) was added for 10 min, then 10 μM nicotine was added for 10 min, followed by the addition of 100 ng/ml LPS and a 4-h incubation. After the incubation period, cells were pelleted, the supernatant removed, and the pellet frozen at −80 oC until RNA was extracted. A measurable TNFα response to LPS stimulation was used as an internal control.
Results
In all of the lymphoblastoid cell lines studied, the microarray signal intensities from genes in the 15q13.3 region were below detection sensitivity except for KLF13 and MTMR10, both of which had detectable levels in all samples except for the homozygous deletion. It is not uncommon for microarrays to lack the sensitivity to detect some genes, especially those involved in signaling, which may only require a few copies of the mRNA. As CHRNA7 and TRPM1 were of greatest interest, we analyzed them using QRT-PCR. We detected signals from both genes from all subjects and the brain tissue, with the exception of the proband (Figures 1 and 2). Although both mRNAs were detected in the lymphoblasts, their levels were much higher in the brain tissue (Supplementary Table 1b). The detection of each gene (CT) in the three subjects with heterozygous deletions (mother, father, and paternal grandmother) was approximately one cycle later than age- and sex-matched comparison subjects (Supplementary Tables 1a and 1b, and Figures 1 and 2), indicating these individuals had approximately 50% less mRNA from these two genes than the comparison subjects. No detectable signal was observed at 50 PCR cycles for any of the genes in the 15q13.3 region in the proband (Supplementary Table 1).
Analysis of the array data, using relatively stringent filtering criteria, identified 267 genes with significantly altered expression (50 genes with reduced expression and 217 with increased expression) in lymphoblastoid cells from the proband with a homozygous deletion of 15q13.3 compared with lymphoblastoid cells from three typically developing comparison male subjects (a complete list of these data are presented in Supplementary Table 2). Of these 267 genes, 28 were also significantly changed in the proband’s father (24 changed in the same direction) and 11 were significantly changed in expression in the proband’s mother (8 changed in the same direction), relative to appropriate age- and sex-matched comparison subjects. However, none of the genes were significantly changed in the proband and both the parents. The paternal grandmother with heterozygous deletion also had 11 of the 267 genes with significantly changed expression, one shared with the father and three different genes shared with the mother.
We used IPA to assess the nature of the genes with differential expression in the proband. Figure 3 indicates direct associations known to exist between the genes located in 15q13.3 and genes found to be differentially expressed in the cell lines. Four genes were of particular interest, because they are known to influence brain and muscle development, (MAPK3(ERK1), MAPK1(ERK2), STAT4, and FASLG; Figure 3 and also see Supplementary Table 3 for explanation of the IPA diagram). MAPK3(ERK1), MAPK1(ERK2), STAT4, and FASLG are all overexpressed in the lymphoblasts with the homozygous deletion of 15q13.3. These genes have been associated with encephalopathy,28, 29 and are intermediaries in the neuregulin and relaxin signaling.
It is interesting that all four of these genes are involved in inflammatory responses and interact with TNFα. Wang et al27 showed that when activated, α7nAChR blocks the induction of TNFα by LPS, thus modulating the inflammatory response. We speculated that the homozygous deletion of α7nAChR might result in an increase basal rate of TNFα. However, we did not see a significant change in the expression of TNFα in the lymphoblastoid cells lacking the CHRNA7 gene. We then exposed the lymphoblastoid cells from two typically developing boys and the proband (with a 15q13.3 homozygous deletion) to LPS, to determine what effect the loss of α7nAChR would have on TNFα response to LPS. All cell lines had an increase in TNFα in response to incubation with LPS (Figure 4). Nicotine reduced the response to LPS in the cells from the typically developing males, but appeared to enhance the LPS response in the cells that had the homozygous deletion. Furthermore α-btx, which competitively binds to α7nAChR with high affinity, prevented the ability of nicotine to inhibit the response to LPS in the typically developing cells, but did not have a significant effect on the cells lines with the CHRNA7 homozygous deletion (Figure 4).
Discussion
The 15q13.3 heterozygous microdeletions have been identified in apparently healthy persons as well as in individuals who have a broad spectrum of neurodevelopmental and neuropsychiatric disorders.1, 2, 3, 4, 5, 6, 7 To date, all individuals with the homozygous 15q13.3 deletion are reported to have a similar and much more severe developmental encephalopathy and neuromuscular disorder than those with the heterozygous deletion.13, 14, 15, 16, 17 We hypothesized that the 15q13.3 deletion had detectable effects on downstream gene expression and the effects of downstream gene dysfunction might contribute to the syndrome.
We compared mRNA expression levels in lymphoblastoid cell lines and brain homogenate samples. We found that the message levels of CHRNA7 and TRPM1 in the brain homogenates were two- to fivefold higher than in the control lymphoblastoid cell lines (Supplementary Table 1b), perhaps explaining why the signal from lymphoblastoid cells was not strong enough for detection by microarray. Much higher expression of CHRNA7 and TRPM1 in the brain highlights the potential importance of these genes in the central nervous system. Nevertheless, we showed that gene expression is detectable in lymphoblastoid cell lines by QRT–PCR, and that expression levels are dose dependent (ie, reduced by approximately 50% in cells with heterozygous deletions as assessed by QRT–PCR). This result validates the cell lines as a useful experimental model, at least as an approximation of relative expression levels of these genes in the central nervous system (CNS).
Analysis of the expression microarray yielded 267 genes with significantly altered expression levels (Supplementary Table 2). The IPA analysis showed 4 of the 267 genes with known direct or indirect association with 3 of the 15q13.3 deleted genes (the other 4 genes in the deleted region returned no known interactions with the genes differentially expressed in the cell lines). Lumican (LUM) was one of the genes differentially expressed in the expression array (Figure 3). However, Lum has no known direct association with TRPM1 other than the fact that both genes affect vision. LUM is a proteoglycan present in the cornea, connective tissues, and heart and skeletal muscle.30 A zebrafish LUM knockout model resulted in scleral thinning and ocular enlargement, among other changes.31 Our patient has a markedly abnormal ERG with a depressed b-wave, a finding consistent with loss of TRPM1 and CSNB.18, 19, 20, 21 It is possible that the differential expression of LUM in the proband may be secondary to an indirect effect of CSNB.
TNFα has been shown to affect synaptic plasticity by regulating synaptic scaling.23 In addition, TNFα as has been shown to be modulated by α7nAChR activation.27 CHRNA7 encodes the α7nAChR, which forms a homomeric, pentameric ligand-gated ion channel highly permeable to calcium.32 The α7nAChR has been implicated in multiple neurocognitive functions as well as epilepsy.33 TNFα secretion is directly inhibited by stimulation of α7nAChRs both in the CNS and through the inflammatory reflex of cytokine-producing macrophages.34 Shen and Yakel35 reported the presence of functional α7nAChRs on the astrocytes of rat hippocampal slices. In patients with a complete loss of CHRNA7, the lack of α7nAChRs might result in increased secretion of TNFα, upregulation of AMPA receptors, and a hyperexcitable state affecting both cognition (through altered synaptic scaling) and epilepsy. However, in our experimental cell model there was no significant difference in the basal rate of TNFα production. We stimulated TNFα production by submitting the cells to a 4-h incubation in LPS. We speculated that LPS activation of TNFα production should be inhibited by nicotine (an agonist of nicotinic receptors) through stimulation of the α7nAChRs. We observed a robust response in TNFα production after LPS incubation and an inhibition of this response by nicotine in cells with a full complement of CHRNA7. The inhibitory effect of nicotine was lost in the cell line lacking CHRNA7, conversely producing a potentiating response following nicotine exposure (Figure 4). To confirm α7nAChR activation, we blocked the receptors by pre-incubation with α-btx, a highly selective competitive antagonist of the α7 nicotinic receptor. As expected, the modulatory effect of nicotine on TNFα production was blocked in the controls, but had no effect in the cells lacking CHRNA7 (Figure 4). These observations are consistent with those of Wang et al.27 Although our assessment was limited to a small number of cell lines with some variability in LPS and nicotine treatment response, these data suggest the obvious influence of α7nAChR on TNFα expression. These observations are consistent with a potential mechanism linking the lack of the α7 neuronal nicotinic activation with impaired synaptic scaling through TNFα dysregulation. The resultant hyperexcitability and impaired synaptic scaling may in turn predispose these patients to developing epilepsy and cognitive impairments.
We identified four genes that were differentially expressed in the proband that had known direct associations with one or more of the genes deleted in 15q13.3: STAT4, FASLG, and ERK1 and ERK2 (Figure 3). STAT4 is a member of the signal transducer and activator of transcription family.36 A murine knockout model of Stat4 showed impaired IL12-mediated functions.37 IL12 is known to be an activator of nitric oxide synthase (NOS), which is important for CNS nitric oxide modulation and synaptic plasticity.38 In the Stat4 knockout mouse, NOS1 protein expression levels were significantly increased. In this study, STAT4 was differentially expressed and the role of dysregulated STAT4/IL12/NOS in the 15q13.3 deletion phenotypes is of great interest.17
STAT4 may also be an important modulator of normal muscle function. Dogra et al39 showed that STAT4 is significantly increased in the muscle of a mouse model of Duchenne muscular dystrophy. It is quite striking that severe hypotonia is reported in all of the patients with homozygous 15q13.3 microdeletions, including the two patients with a deletion limited to CHRNA7.13, 17 Therefore, it is possible that STAT4 may contribute to the profound hypotonia noted in patients with 15q13.3 homozygous deletion syndrome. We cannot exclude a contribution of the other genes, such as MTMR10, included in the deleted region.15 However, the observations that at least one child with a deletion limited to CHRNA7 is hypotonic, and that in our expression array none of the 267 differentially expressed genes had any currently known direct or indirect interaction with MTMR10, make it somewhat less likely that MTMR10 has a significant role in the presently observed phenotype.
FASLG (TNFSF6) encodes a widely expressed transmembrane protein that is important for initiating apoptosis. It is part of the TNF superfamily and participates in initiating cell activation and cell differentiation to promote chronic inflammation and inflammatory responses.40 Upregulation of FASLG in the CNS has been associated with neurons and oligodendrocytes undergoing Fas-mediated apoptosis, resulting in neurodegeneration and demyelination, respectively (reviewed in Choi and Benveniste41).
Extracellular signal-regulated kinases, ERK1 and ERK2, are part of the MAPK signal cascade, which is essential for signal transduction in the CNS. ERK1 and ERK2 have been found to have important roles in several neurological syndromes, including neurocardio–facial–cutaneous syndromes.42 Newbern et al43 created a conditional Erk1/2 mouse knockout and showed that Erk1/2 deletion resulted in massive hypomyelination. It is possible that through dysregulation of the MAPK/ERK pathway, both cognitive function and muscle tone may be affected. This may represent another pathway ultimately contributing both to the profound cognitive impairment and the hypotonia observed in patients with the homozygous 15q13.3 deletions.
One of the challenges of interpretation of the results of comparative genomic hybridization microarrays is to understand how the observed deletions and duplications cause cellular dysfunction. This is especially challenging, as patients with similar genomic abnormalities often have a quite variable phenotypic presentation. Girirajan and Eichler44 proposed a ‘two hit’ mechanism in which phenotype is greatly modified in the presence of a secondary genetic/genomic disturbance. We hypothesize that the phenotype of patients with heterozygous deletions will be modified by a ‘second hit,’ possibly involving one of the genes we were able to identify in this study.
One of the limitations of this study is that our analysis was limited to a single patient with the homozygous deletion. We used a local error model to assess for statistical significance in the reported levels of expression. As more patients with these deletions become identified, and as their cell lines become available from central repositories, we anticipate that these results will be repeated. Thus, our study offers some potentially important avenues for closer scrutiny. Another intrinsic limitation was that we used lymphoblastoid cell lines as our expression assay. It is probable that these cells do not perfectly model neuronal cell function. Nevertheless, the relative expression levels of CHRNA7 and TRMP1 accurately reflected dosage changes between the control subjects and our subjects with heterozygous deletions. Furthermore, many of the affected genes are ubiquitously expressed. Therefore, it is reasonable to assume that the differential expression in the cell lines is a reasonable approximation of gene expression in the CNS.
In conclusion, using immortalized lymphoblastoid cell lines, we modeled differential gene expression from a patient with a 15q13.3 homozygous deletion. Of the seven deleted genes in the proband, four were found to have known relationships to genes that were differentially expressed in our model system. The functional significance of these pathways is consistent with the previous suggestion that the CHRNA7 deletion may contribute to much of the observed phenotype in patients with homozygous deletions. We further tested this hypothesis by showing that TNFα production is modulated by α7nAChRs. The lack of modulation in patients with homozygous deletions of CHRNA7 may result in hyperexcitability and impaired synaptic scaling. We further propose that, according to the second hit model, the variable phenotypic presentations of at least some of the patients with heterozygous deletions may be explained by mutations in the genes uncovered in this study.
Accession codes
References
Mulley JC, Dibbens LM : Chipping away at the common epilepsies with complex genetics: the 15q13.3 microdeletion shows the way. Genome Med 2009; 1: 33.
Ben-Shachar S, Lanpher B, German JR et al: Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet 2009; 46: 382–388.
Pagnamenta AT, Wing K, Akha ES et al: A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet 2009; 17: 687–692.
Miller DT, Shen Y, Weiss LA et al: Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet 2009; 46: 242–248.
Sharp AJ, Mefford HC, Li K et al: A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 2008; 40: 322–328.
Helbig I, Mefford HC, Sharp AJ et al: 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet 2009; 41: 160–162.
van Bon BW, Mefford HC, Menten B et al: Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet 2009; 46: 511–523.
Shinawi M, Schaaf CP, Bhatt SS et al: A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet 2009; 41: 1269–1271.
Gault J, Robinson M, Berger R et al: Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7). Genomics 1998; 52: 173–185.
Elmslie FV, Rees M, Williamson MP et al: Genetic mapping of a major susceptibility locus for juvenile myoclonic epilepsy on chromosome 15q. Hum Mol Genet 1997; 6: 1329–1334.
Neubauer BA, Fiedler B, Himmelein B et al: Centrotemporal spikes in families with rolandic epilepsy: linkage to chromosome 15q14. Neurology 1998; 51: 1608–1612.
Martin LF, Leonard S, Hall MH, Tregellas JR, Freedman R, Olincy A : Sensory gating and alpha-7 nicotinic receptor gene allelic variants in schizoaffective disorder, bipolar type. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 611–614.
Hoppman-Chaney N, Wain K, Seger PR, Superneau DW, Hodge JC : Identification of single gene deletions at 15q13.3: further evidence that CHRNA7 causes the 15q13.3 microdeletion syndrome phenotype. Clin Genet 2012, e-pub ahead of print 9 July 2012.
Masurel-Paulet A, Andrieux J, Callier P et al: Delineation of 15q13.3 microdeletions. Clin Genet 2010; 78: 149–161.
LePichon JB, Bittel DC, Graf WD, Yu S : A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7, and other homozygously deleted genes. Am J Med Genet A 2010; 152A: 1300–1304.
Endris V, Hackmann K, Neuhann TM et al: Homozygous loss of CHRNA7 on chromosome 15q13.3 causes severe encephalopathy with seizures and hypotonia. Am J Med Genet A 2010; 152A: 2908–2911.
Liao J, DeWard SJ, Madan-Khetarpal S, Surti U, Hu J : A small homozygous microdeletion of 15q13.3 including the CHRNA7 gene in a girl with a spectrum of severe neurodevelopmental features. Am J Med Genet A 2011; 155A: 2795–2800.
Audo I, Kohl S, Leroy BP et al: TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet 2009; 85: 720–729.
Li Z, Sergouniotis PI, Michaelides M et al: Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet 2009; 85: 711–719.
Nakamura M, Sanuki R, Yasuma TR et al: TRPM1 mutations are associated with the complete form of congenital stationary night blindness. Mol Vis 2010; 16: 425–437.
van Genderen MM, Bijveld MM, Claassen YB et al: Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet 2009; 85: 730–736.
Webber C : Functional enrichment analysis with structural variants: pitfalls and strategies. Cytogenet Genome Res 2011; 135: 277–285.
Stellwagen D, Malenka RC : Synaptic scaling mediated by glial TNF-alpha. Nature 2006; 440: 1054–1059.
Bittel DC, Kibiryeva N, Sell SM, Strong TV, Butler MG : Whole genome microarray analysis of gene expression in Prader-Willi syndrome. Am J Med Genet A 2007; 143: 430–442.
Bittel DC, Kibiryeva N, McNulty SG, Driscoll DJ, Butler MG, White RA : Whole genome microarray analysis of gene expression in an imprinting center deletion mouse model of Prader-Willi syndrome. Am J Med Genet A 2007; 143: 422–429.
Eisenhart C : The assumptions underlying the analysis of variance. Biometrics 1947; 3: 1–21.
Wang H, Yu M, Ochani M et al: Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature 2003; 421: 384–388.
Matsukawa A : STAT proteins in innate immunity during sepsis: lessons from gene knockout mice. Acta Med Okayama 2007; 61: 239–245.
Lee ST, Chu K, Sinn DI et al: Erythropoietin reduces perihematomal inflammation and cell death with eNOS and STAT3 activations in experimental intracerebral hemorrhage. J Neurochem 2006; 96: 1728–1739.
Chakravarti S, Magnuson T : Localization of mouse lumican (keratan sulfate proteoglycan) to distal chromosome 10. Mamm Genome 1995; 6: 367–368.
Yeh LK, Liu CY, Kao WW et al: Knockdown of zebrafish lumican gene (zlum) causes scleral thinning and increased size of scleral coats. J Biol Chem 2010; 285: 28141–28155.
Shen JX, Yakel JL : Nicotinic acetylcholine receptor-mediated calcium signaling in the nervous system. Acta Pharmacol Sin 2009; 30: 673–680.
Russo P, Taly A : α7-Nicotinic acetylcholine receptors: an old actor for new different roles. Curr Drug Targets 2012; 13: 574–578.
Rosas-Ballina M, Olofsson PS, Ochani M et al: Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science 2011; 334: 98–101.
Shen JX, Yakel JL : Functional alpha7 nicotinic ACh receptors on astrocytes in rat hippocampal CA1 slices. J Mol Neurosci 2012; 48: 14–21.
Diefenbach A, Schindler H, Rollinghoff M, Yokoyama WM, Bogdan C : Requirement for type 2 NO synthase for IL-12 signaling in innate immunity. Science 1999; 284: 951–955.
Hong MS, Song JY, Yun DH, Cho JJ, Chung JH : Increase of NADPH-diaphorase expression in hypothalamus of stat4 knockout mice. Korean J Physiol Pharmacol 2009; 13: 337–341.
Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM : Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci 2007; 8: 766–775.
Dogra C, Srivastava DS, Kumar A : Protein-DNA array-based identification of transcription factor activities differentially regulated in skeletal muscle of normal and dystrophin-deficient mdx mice. Mol Cell Biochem 2008; 312: 17–24.
Ma Y, Liu H, Tu-Rapp H et al: Fas ligation on macrophages enhances IL-1R1-Toll-like receptor 4 signaling and promotes chronic inflammation. Nat Immunol 2004; 5: 380–387.
Choi C, Benveniste EN : Fas ligand/Fas system in the brain: regulator of immune and apoptotic responses. Brain Res Brain Res Rev 2004; 44: 65–81.
Bentires-Alj M, Kontaridis MI, Neel BG : Stops along the RAS pathway in human genetic disease. Nat Med 2006; 12: 283–285.
Newbern JM, Li X, Shoemaker SE et al: Specific functions for ERK/MAPK signaling during PNS development. Neuron 2011; 69: 91–105.
Girirajan S, Eichler EE : Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet 2010; 19: R176–R187.
Acknowledgements
We thank the patient and his family for their cooperation. This research was supported by a generous endowment from the State of Kansas Fraternal order of Eagles.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Le Pichon, JB., Yu, S., Kibiryeva, N. et al. Genome-wide gene expression in a patient with 15q13.3 homozygous microdeletion syndrome. Eur J Hum Genet 21, 1093–1099 (2013). https://doi.org/10.1038/ejhg.2013.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.1
Keywords
This article is cited by
-
Local and global chromatin interactions are altered by large genomic deletions associated with human brain development
Nature Communications (2018)
-
Modulatory effects of α7 nAChRs on the immune system and its relevance for CNS disorders
Cellular and Molecular Life Sciences (2016)