Abstract
Wnt signalling is one of a few pathways that are crucial for controlling genetic programs during embryonic development as well as in adult tissues. WNT10A is expressed in the skin and epidermis and it has shown to be critical for the development of ectodermal appendages. A nonsense mutation in WNT10A was recently identified in odonto-onycho-dermal dysplasia (OODD; MIM 257980), a rare syndrome characterised by severe hypodontia, nail dystrophy, smooth tongue, dry skin, keratoderma and hyperhydrosis of palms and soles. We identified a large consanguineous Pakistani pedigree comprising six individuals affected by a complete OODD syndrome. Autozygosity mapping using SNP array analysis showed that the affected individuals are homozygous for the WNT10A gene region. Subsequent mutation screening showed a homozygous c.392C>T transition in exon 3 of WNT10A, which predicts a p.A131V substitution in a conserved α-helix domain. We report here on the first inherited missense mutation in WNT10A with associated ectodermal features.
Similar content being viewed by others
Introduction
Wnt signalling is a highly conserved pathway across species ranging from worms, flies to vertebrates.1, 2, 3 Mouse models targeted for different Wnt genes are lethal or display severe developmental defects illustrating the importance of Wnt signalling during development.4, 5, 6 The signalling is initiated by one of 19 WNT ligands that bind to and activate specific receptors, such as the frizzled and LRP families. The downstream transduction comprises the canonical WNT-β-catenin pathway and the non-canonical branches.6, 7 The critical functions of WNT proteins during development are illustrated by heterozygous mutations in the gene encoding PORCN, predicted to be involved in palmitoylation and secretion of WNT. PORCN mutations are associated with the multisystem disease, focal dermal hypoplasia,8 involving organs of both mesodermal and ectodermal origin.
Several members of the WNT family are expressed in the skin and during the formation of ectodermal appendages, such as hair-follicles and teeth.9, 10, 11, 12 WNT10A and WNT10B are highly expressed in embryonic skin and in the placodes at follicle morphogenesis,13 and WNT10A seems to be critical for dentinogenesis and tooth morphogenesis in regulating the odontoblast differentiation.14 It was recently shown that a WNT10A nonsense mutation is associated with the rare ectodermal dysplasia subtype odonto-onycho-dermal dysplasia or tricho-odonto-onycho-dermal dysplasia (OODD).15, 16, 17, 18, 19 This observation suggests that WNT10A, in addition to the formation of teeth and hair follicles, is of importance for the formation of nails, regeneration of the epidermis, papillae of the tongue and sweat gland function. The nonsense mutation predicts a complete loss of WNT10A function and a disrupted WNT10A signalling. This raises the question whether missense mutations in WNT10A may result in a partial OODD phenotype. We show herein that a missense mutation in a highly conserved WNT10A domain is associated with developmental defects and a complete OODD syndrome.
Materials and methods
Patients
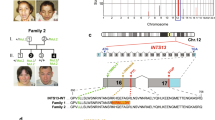
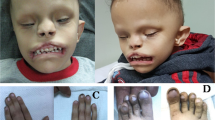
We identified and investigated a large consanguineous Pakistani family. The family comprises six individuals affected by tricho-odonto-onycho-dermal dysplasia (Figure 1a). The inheritance pattern is autosomal recessive. Both the father and three of his children are affected in one loop of the family illustrating the high degree of consanguinity over several generations. A total of six members (IV:3, V:2, V:3,V:5, V:6 and V:7) were examined and the clinical findings are given in Table 1. Affected individuals presented with thin eyelashes, sparse scalp and body hair. The skin was dry with pilar keratosis and diffuse follicular hyperkeratosis. Keratoderma and palmar erythema was also noticed in affected individuals (Figure 1b). Scalp hair was reported absent in two patients at birth. Affected individuals complained of hyperhidrosis in palms and soles as well as heat intolerance. Nail plates of fingers were thin and toe-nail dysplasia was present in two patients (Figure 1c). Oral examination showed severe hypodontia with few permanent and abnormally shaped teeth, as well as a marked reduction or absence of mainly filiform papillae of the tongue having a smooth appearance (Figure 1d). In the absence of permanent teeth some deciduous teeth remained into adulthood (Figure 1e).
Pedigree segregating OODD and clinical features. (a) Several consanguinity loops are shown and the OODD phenotypes appear in the two successive generations IV and V (right loop). Filled symbols denote affected individuals. (b) Sole of right foot in individual V:7 at age 37 years illustrating plantar keratoderma. (c) Dystrophic toe-nails in individual V:2 at age 48 years. (d) Smooth tongue with reduced papillae in individual V:6 at 23 years of age. (e) Panoramic radiograph of the affected male V:3 at 39 years of age showing agenesis of most permanent teeth, third molars excluded. In the upper jaw, there are two permanent central incisors and one permanent premolar with conical crown shape (upper arrows). In the lower jaw, there are four permanent premolars with reduced crown form (bottom arrows).
Genotyping
Venous blood samples were obtained from five healthy and six affected family members. Genomic DNA was extracted from lymphocytes according to the standard procedures. SNP genotyping was performed on genomic DNA from four affected family members (V:2, V:3, V:5 and V:7) using the GeneChip Mapping 250K array (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's protocol. Homozygosity mapping and sorting of genomic regions were performed with the dedicated software, AutoSNPa (University of Leeds, Leeds, UK). A cutoff of >30 homozygous SNPs was used for the selection of candidate regions.20 Selected regions were further investigated by genotyping all family members with microsatellite markers derived from known and novel polymorphic repeats (primer sequences are available upon request). Two-point LOD scores were calculated using the MLINK program of FASTLINK (Human Genome Mapping Project Resources Centre, Cambridge, UK) computer package21 assuming an autosomal recessive inheritance, full penetrance and a disease allele frequence of 10−5. The pedigree was drawn with the Cyrillic software, version 2.1.3 (Cherwell Scientific Publishing Ltd, Oxford, UK) and the haplotype analysis was performed manually.
Sequencing
All exons of the WNT10A gene including the 5′-UTR and intron–exon boundaries were analysed on genomic DNA from two affected family members. Each PCR reaction was performed on 50 ng genomic DNA in the presence of 0.025 U/μl Platinum Taq Polymerase (Invitrogen, Calsbad, CA, USA), 1.5 mM MgCl2, 200 mM dNTPs using 0.2 μ M intronic primers in 20 μl reaction volume (primer sequences are available upon request). The sequencing reactions were generated using the BigDye Terminator v3.1 Cycle Sequencing Kit (Invitrogen, San Diego, CA, USA) according to manufacturer's protocol and separated on an ABI 3700 (Applied Biosystems, Foster City, CA, USA). The c.392C>T transition was analysed on all 11 sampled family members and on 800 control chromosomes (400 of Pakistani origin and 400 of Swedish origin).
Bioinformatic analyses
We used the National Center for Biotechnology Information (NCBI) Entrez Genome Map Viewer and Ensemble Human Genome Server Database for the localisation of microsatellite marker loci. The genome database was used for information on microsatellite markers and their primer sequences. The WNT10A sequence is based on GenBank accession number NM_025216.2. Sequences for human WNT10A protein and its orthologs (ie, Pan troglodytes, Canis lupus familiaris, Bos Taurus, Mus musculus, Rattus norvegicus, Danio rerio, Gallus gallus, Drosophila melanogaster and Anopheles gambiae) were retrieved from HomoloGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene) and subsequently aligned using BioEdit v7.0.5.3 (Ibis Biosciences, Carlsbad, CA, USA).22 The secondary structure of Human WNT10A was analysed using PSIPRED (University College of London, London, UK).23, 24
Results
Linkage to the five gene regions associated with different forms of EDs was excluded using microsatellite markers. We then obtained genotypes from the 250K SNP array. We identified five candidate regions that seemed to be homozygous for >30 adjacent SNPs in samples from four affected individuals (V:2, V:3, V:5 and V:7; Figure 1a). Linkage was confirmed only to the chromosome 2 region using multiallelic microsatellite markers on samples from parents and healthy siblings with a maximum two-point LOD score of 2.97 obtained for marker locus D2S434 (θ=0.00). The gene region coincides with the position of WNT10A and the earlier association to OODD prompted us to screen this gene for mutations. Sequencing showed a homozygous c.392C>T transition in all six affected family members (Figure 2). The mutation is located in exon 3 and results in a p.A131V substitution. This missense variant was excluded on 400 control chromosomes of Pakistani origin and 400 control chromosomes of Swedish origin.
Sequence chromatograms of part of WNT10A showing the position corresponding to c.392C and flanking sequences. (a) Chromatogram from a healthy control individual. (b) Chromatogram from individual IV:4 in the pedigree illustrating heterozygosity for c.392C>T and the normal sequence. (c) Chromatogram from individual V:5 affected by OODD showing homozygosity for the c.392C>T transition.
The effect of the p.A131V mutation on protein structure and function was analysed by comparing the human wild-type WNT10A protein sequence to orthologs from a series of animals. The results indicate that the human A131 residue is highly conserved across species (Figure 3). Furthermore, we used the primary protein sequence to predict the secondary structure of human WNT10A. According to the analysis, human WNT10A is a protein of 46.4 kDa containing 10 putative α-helices and seven putative β-strands (Figure 3). The A131 residue is positioned at the N-terminus of helix 4.
Schematic representation of structural secondary elements of the human WNT10A protein as predicted by PSIPRED. The predicted elements (PRED) are indicted above the amino acid (AA) sequence (coils: straight line; helices: grey cylinders numbered h1–h11; strands: black arrows numbered s1–s7). The boxed area illustrates human AA 121–160 with multiple species alignment for the region flanking the p.A131V mutation indicated in grey. Numbers to the left of the alignment refer to the first amino acid counted from the N-terminus of each WNT10A ortholog (H.s., Homo sapiens; P.t., Pan troglodytes; C.f., Canis lupus familiaris, B.t., Bos Taurus; M.m., Mus musculus; R.n., Rattus norvegicus; D.r., Danio rerio; G.g., Gallus gallus; D.m. Drosophila melanogaster; A.g., Anopheles gambiae).
Discussion
We present herein a homozygous WNT10A missense mutation c.392C>T in four cases of the OODD syndrome. This is the first reported missense mutation in WNT10A and results in a p.A131V substitution. The patients belong to a single consanguineous Pakistani family comprising six living affected family members. A WNT10A nonsense mutation (c.697G>T; p.E233X) was earlier reported in six distantly related OODD patients.19 With some clinical variabilities, these patients showed hypodontia, dystrophic nails, palmoplantar keratoderma and hyperhidrosis of palms and sole, abnormal hair pattern, dry skin and smooth tongue with absent or reduced papillae. All six patients in our study have severe hypodontia, smooth tongue with reduced papillae, hyperhidrosis of palms and soles, diffuse follicular hyperkeratosis and dry skin. Several patients also show palmoplantar keratoderma (5/6), abnormal hair pattern (5/6) and dystrophic toe-nails (5/6). However, differences were noted in the formation of nails when comparing our OODD patients with the earlier described OODD phenotype associated with a WNT10A nonsense mutation. None of our patients were reported with congenital absence of nails. Moreover, we observed fingernail dystrophy in only one out of six affected family members. In the earlier report, the WNT10A nonsense mutation was associated with total absence of nails in some individuals and dystrophic fingernails in five out of six affected individuals. Apart from these differences, there are striking similarities when comparing the two OODD phenotypes associated with a missense mutation and a nonsense mutation. Consequently, our findings suggest that the p.A131V substitution has a deleterious effect on WNT10A signalling, possibly through a loss-of-function mechanism.
The WNT genes encode a large family of secreted glycoproteins that specify various cell lineages during embryogenesis.3, 4 The WNT signals are transduced through a canonical pathway for cell fate determination and through a non-canonical pathway for control of cell movement and tissue polarity.6, 7, 25, 26 It is yet unclear, whether WNT10A mediates its function through one or both of these pathways. The p.A131V substitution presented here affects a residue and a domain that is highly conserved. This suggests that the A131 residue is functionally or structurally important. Other domains of the protein show a considerably lower degree of conservation. The p.A131 residue is not located in the structural elements of WNT proteins, such as the WNT family signature (amino acids 262–271) or the 24 conserved cysteine residues that are believed to be important for intramolecular disulphide bridge formations.2, 23 To clarify whether the mutation designates any other region of structural importance of WNT10A, we performed the secondary structure prediction. According to the analysis, the p.A131 residue is positioned within the N-terminal side of helix 4. The substitution may possibly alter the length or stability of the helix and, consequently, the three-dimensional folding of the protein. Yet another possibility is that the A131 residue is involved in interactions with downstream proteins in the signalling pathway (eg, frizzled receptors). The substitution replaces a short alanine with a bulkier valine that could possibly affect the WNT10A signal transduction.
In summary, we provide further evidence of WNT10A mutations in OODD. We show that a homozygous missense mutation is associated with a complete OODD phenotype contributing to the classification of ectodermal dysplasias.27, 28 The findings underscore the importance of WNT10A in the formation of ectodermal appendages, such as hair, teeth, skin and nails in humans. The identification of human phenotypes with other WNT10A mutations, or possibly mutations in downstream members of this signalling pathway, will be important to further understand developmental processes of the ectoderm.
Accession codes
References
Nelson W, Nusse R : Convergence of Wnt, beta catenin and cadherin pathways. Science 2004; 303: 1483–1487.
Coudreuse D, Korswagen HC : The making of Wnt: new insights into Wnt maturation, sorting and secretion. Development 2007; 134: 3–12.
Nusse R : Wnt signaling and stem cell control. Cell Res 2008; 18: 523–527.
Logan CY, Nusse R : The Wnt signaling pathway in development and disease. Ann Rev Dev Biol 2004; 20: 781–810.
Van Amerongen R, Berns A : Knock-out mouse models to study Wnt signal transduction. Trends Genet 2006; 22: 678–689.
Clevers H : Wnt/beta-catenin signalling in development and disease. Cell 2006; 127: 469–480.
Moon RT, Kohn AD, De Ferrari GV, Kaykas A : WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet 2004; 5: 691–701.
Grzeschik KH, Bornholdt D, Oeffner F et al: Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat Genet 2007; 39: 820–821.
Wang J, Shackleford GM : Murine Wnt10a and Wnt10b: cloning and expression in developing limps, face and skin of embryos and in adults. Oncogene 1996; 13: 1537–1544.
Dassule HR, McMahon AP : Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol 1998; 20: 215–227.
Millar SE, Willert K, Salinas PC et al: WNT signaling in the control of hair growth and structure. Dev Biol 1999; 207: 133–149.
Andl T, Reddy ST, Gaddapara T, Millar SE : WNT signals are required for the initiation of hair follicle development. Dev Cell 2002; 2: 643–653.
Reddy S, Andl T, Bagasra A et al: Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech Dev 2001; 107: 69–82.
Yamashiro T, Zheng L, Shitaku Y et al: Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation 2007; 75: 452–462.
Pinheiro M, Pereira LC, Freire-Maia N : A previously undescribed condition: tricho-odonto-onycho-dermal syndrome. A review of the tricho-odonto-onychial subgroup of ectodermal dysplasias. Br J Dermatol 1981; 105: 371–382.
Fadhil M, Ghabra TA, Deeb M, Der Kaloustian VM : Odontoonychodermal dysplasia: a previously apparently undescribed ectodermal dysplasia. Am J Med Genet 1983; 14: 335–346.
Mégarbané A, Noujeim Z, Fabre M, Der Kaloustian VM : New form of hidrotic ectodermal dysplasia in a Lebanese family. Am J Med Genet 1998; 75: 196–199.
Megarbané H, Haddad M, Delague V, Renoux J, Boehm N, Megarbane A : Further delineation of the odonto-onycho-dermal dysplasia syndrome. Am J Med Genet A 2004; 129A: 193–197.
Adaimy L, Chouery E, Megarbané H et al: Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am J Hum Genet 2007; 81: 821–828.
Dahlqvist J, Klar J, Hausser I et al: Congenital Ichthyosis: mutations in ichthyin associated with specific structural abnormalities in the granular layer of epidermis. J Med Genet 2007; 44: 615–620.
Cottingham R, Indury RM, Schaffer AA : Faster sequential genetic linkage computations. Am J Hum Genet 1993; 53: 252–263.
Hall T : BioEdit: a user friendly biological sequence alignment editor and analysis programme for Windows. Nucl Acids Symp Ser 1999; 41: 95–98.
Jones DT : Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 1999; 292: 195–202.
McGuffin LJ, Bryson K, Jones DT : The PSIPRED protein structure prediction server. Bioinformatics 2000; 16: 404–405.
Zeng X, Huang H, Tamai K et al: Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development 2008; 135: 367–375.
Klaus A, Birchmeier W : Wnt signalling and its impact on development and cancer. Nat Rev Cancer 2008; 8: 387–398.
Priolo M, Lagana C : Ctodermal dysplasias: a new clinical-genetic classification. J Med Genet 2001; 38: 579–585.
Lamartine J : Towards a new classification of ectodermal dysplasias. Clin Exp Dermatol 2003; 28: 351–355.
Acknowledgements
We thank the families and patients that participated in this study. We are also indepted to Lynn Adaimy for providing WNT10A primer sequences. This study was supported by The Swedish Research Council, Swedish Links (Asia), The Indevelops fund, Thuréus fund, Torsten and Ragnar Söderbergs Funds, Uppsala University and the Uppsala University Hospital.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nawaz, S., Klar, J., Wajid, M. et al. WNT10A missense mutation associated with a complete Odonto-Onycho-Dermal Dysplasia syndrome. Eur J Hum Genet 17, 1600–1605 (2009). https://doi.org/10.1038/ejhg.2009.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.81
Keywords
This article is cited by
-
WNT10A variants: following the pattern of inheritance in tooth agenesis and self-reported family history of cancer
Clinical Oral Investigations (2022)
-
Distinct tooth regeneration systems deploy a conserved battery of genes
EvoDevo (2021)
-
Prevalence of WNT10A gene mutations in non-syndromic oligodontia
Clinical Oral Investigations (2019)
-
WNT10A variants in relation to nonsyndromic hypodontia in eastern Slovak population
Journal of Genetics (2018)
-
The association between WNT10A variants and dental development in patients with isolated oligodontia
European Journal of Human Genetics (2017)