Abstract
Marinesco–Sjögren syndrome (MSS) is a rare autosomal recessively inherited neurodegenerative disorder characterized by cerebellar ataxia, cataracts, mental retardation, and progressive myopathy. Recently, mutations in the SIL1 gene, which encodes an endoplasmic reticulum (ER) resident cochaperone, were identified as a major cause of MSS. We here report four novel mutations in SIL1, including the first missense substitution p.Leu457Pro described in MSS. In addition, we excluded three functional candidate genes, HSPA5, HYOU1, and AARS, as causative genes in SIL1 mutation-negative patients. To understand the mechanisms of disturbed SIL1 function, we studied the subcellular localization of the missense mutant Leu457Pro protein in COS-1 cells. Moreover, we studied a mutant protein lacking the putative C-terminal ER retrieval signal. In contrast to the wild-type protein's localization to ER and Golgi apparatus, both mutant proteins formed aggregates within the ER depending on the expression level. These data imply that aggregation of mutant proteins may contribute to MSS pathogenesis. The genetic background of a subgroup of patients with MSS remains uncovered.
Similar content being viewed by others
Introduction
Marinesco–Sjögren syndrome (MSS, MIM 248800) is an autosomal recessive disorder affecting various tissues. The main clinical findings are cerebellar ataxia with cerebellar atrophy, early-onset cataracts, psychomotor delay, progressive muscular weakness, and muscle atrophy.1, 2, 3 In addition, hypergonadotropic hypogonadism, skeletal abnormalities, short stature, strabismus, hypotonia, dysarthria, and nystagmus are frequent findings. Recently, we4 and others5 reported the main cause for the MSS phenotype to be mutations in the SIL1 gene predicted to result in loss of SIL1 function due to premature termination of translation or abnormal splicing of the transcript. Since the original reports, two other families with mutations in SIL1 have been identified.6, 7 However, some patients with typical MSS do not have identifiable mutations in SIL1, implying genetic heterogeneity.5
SIL1 acts as an adenine nucleotide exchange factor for the heat-shock protein 70 (Hsp70) chaperone GRP78 (also known as BiP and HSPA5), a molecular chaperone functioning mainly in the endoplasmic reticulum (ER).8, 9 SIL1 regulates the ATPase cycle of GRP78 and has thus been anticipated to be involved in protein translocation into the ER, proper folding of the newly synthesized proteins, regulating the degradation of proteins that fail to mature properly, and responding to cell stress.10 However, recent data imply that SIL1 is most likely not involved in the regulation of the unfolded protein response (UPR), as the UPR is upregulated normally in the woozy (wz) mouse, a spontaneous Sil1 mouse mutant.11 SIL1 is a 461 amino-acid protein that has a potential N-terminal ER targeting sequence and a C-terminal tetrapeptide (KELR; amino acids 458–461), quite likely to be an ER retrieval sequence.8 In previous studies, transiently expressed SIL1 has been shown to localize in the ER.8, 11 The tissue expression pattern of SIL1 is similar to that of GRP78 in mouse, thereby supporting their functional interaction.4
The cerebellar atrophy seen in patients with MSS is caused by loss of Purkinje and granule cells.12, 13, 14 The same cerebellar findings are seen in the wz mouse with adult-onset ataxia as the main clinical manifestation.11 Interestingly, both in human patients with MSS and in wz mice, Purkinje cells in cerebellar lobules IX and X, which constitute the vestibulocerebellum, do not degenerate. A similar pattern of Purkinje cell death is seen in another mouse model with an alanyl-tRNA synthetase (Aars) mutation leading to intracellular accumulation of misfolded proteins in neurons.15 The wz mice differ from humans with MSS, since in the mice no symptoms like cataracts or myopathy have been described.11
SIL1 is ubiquitously expressed and it still remains unknown why certain tissues and cell types, for example cerebellum and cerebellar Purkinje cells, are more vulnerable to loss of SIL1 function. A possible explanation is that alternative nucleotide exchange factors for GRP78, such as the ER-resident HYOU1 (also known as ORP150 and GRP170), can compensate for the loss of SIL1 function in certain tissues.4, 16 Alternatively, Purkinje cells may be innately more sensitive to the accumulation of misfolded proteins, as cerebellar Purkinje cell loss and ataxia are prominent findings in many human repeat expansion diseases.17
Here, we screened patients from 23 families for mutations in SIL1 and report four novel mutations. We evaluated the consequence of a novel missense mutation and deletion of the ER retrieval tetrapeptide on the subcellular localization of SIL1 in transfected COS-1 cells. On the basis of information on SIL1 function or the associated phenotypic features in mouse models, we hypothesized that alterations in HSPA5, HYOU1, and AARS might result in MSS phenotype and sequenced these genes in patients negative for SIL1 mutations.
Materials and methods
Subjects and clinical investigations
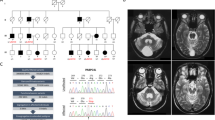
The Institutional Review Board of the Helsinki University Central Hospital, Finland, approved the study. Informed consent was obtained from all subjects and/or their parents/guardians. A DNA sample was available from 26 individuals with a clinical MSS diagnosis in 23 families. The pedigrees of families M7, M13, M20, M21, and M23 are shown in Figure 1. The clinical features of the nine patients with SIL1 mutations are summarized in Table 1.
Pedigrees and MSS-associated SIL1 mutations in five MSS families. DNA samples were available for analysis from the numbered individuals except for patient M705, shown in parenthesis, from whom only clinical information was available. The sequence chromatograms show homozygous SIL1 mutations (underlined in red). (a) Patients M2303 and M2304 are homozygous for a one-base duplication c.936dupG predicted to cause frameshift and premature truncation of translation (p.Leu313AlafsX39). (b) Patient M2003 is homozygous for a T to A transversion c.1367T>A leading to premature termination of translation (p.Leu456X). (c) Patients M2103 and M2104 are homozygous for a T to C transition c.1370T>C leading to substitution of leucine with a cyclic proline (p.Leu457Pro). (d) Patients M704, M1303, and M1304 are homozygous for a G to A transition in intron 9 (c.1030-9G>A), leading to the introduction of a novel splice acceptor site, frameshift, and premature truncation of translation (p.Phe345AlafsX9). The vertical broken lines indicate the exon–intron boundaries in normal control (blue) and affected patient (red) sequences. The similarity of the splice site sequences (underlined in blue in the chromatograms) is illustrated below the chromatogram. (e) The c.1030-9G>A mutation leads to the insertion of seven nucleotides to RNA sequence as shown by RT-PCR analysis.
Patient M704 in family M7 and her sister M705, whose DNA sample was not available for the study, were originally described by Andersen18 as cases 2 and 3 with classical MSS. Family M13 has previously been described elsewhere.1, 19 The affected siblings, M1303 and M1304, show classical MSS with the exception of atrophy of pons detected in magnetic resonance imaging (MRI) in patient M1304. No consanguinity has been reported in families M7 and M13.
Patient M2003 presented at the age of 19 months with developmental delay. He was born prematurely at 31 weeks of gestation. His newborn period was complicated by infant respiratory distress syndrome and recurrent apparent life-threatening events, pulmonary hypertension, and gastroesophageal reflux. Apart from a long philtrum and a thin upper lip, he has no apparent dysmorphic features. His creatine kinase values have been slightly elevated, 283 U/l. His shoulders are fixed in anteroposition, but no other radiological pathology is present. He has a small scrotum. He is prone to hematomas but has normal levels of thrombocytes. His height and head circumference followed −1 SD. His parents are first cousins and are of Turkish origin.
Patients M2103 and M2104 are Japanese brothers. Their parents are second cousins and show no signs of MSS. A twin brother of the proband (M2103) was born preterm and died soon after birth. The affected brothers both showed psychomotor delay and distinct cerebellar symptoms, including limb and truncal ataxia, hypotonia, and dysarthria. They showed poor vision at the early infantile period, which was diagnosed as cataract in their teens and was surgically removed later on. Brain MRI of the brothers showed marked cerebellar atrophy. In addition, M2103 showed shortened metacarpals and metatarsals, and M2104 showed elevated serum creatine kinase.
Patients M2303 and M2304 are Japanese sisters. Their parents are second cousins once removed and their mother has diabetes. The sisters are similarly affected: they both had bilateral cataracts operated at the age of 5 years. They learned to walk alone but their gait was unstable, and they needed the help of a walker in their teenage years. They both lost their ambulation around 20 years of age. The older sister M2303 also has diabetes and osteoporosis.
The other patients participating in this study either showed all the hallmark features of MSS or lacked one or two of the features (cerebellar atrophy and ataxia, cataracts, or myopathy). They also presented with a variable combination of other MSS-associated symptoms and atypical symptoms rarely seen in MSS. The clinical features of the SIL1 mutation-negative patients are summarized in Table 2.
Mutation analysis
We isolated genomic DNA using standard methods. For SIL1 (GenBank NM_001037633), we amplified exons and exon–intron boundaries of the nine coding and the two non-coding exons (1a and 1b) with intronic primers. In SIL1 mutation-negative patients, we amplified the exons and exon–intron boundaries of three functional candidate genes, HSPA5 (GenBank NM_005347), HYOU1 (GenBank NM_006389), and AARS (GenBank NM_001605). The primer sequences are available on request. We purified the amplified products using the PCR Product Pre-Sequencing Kit (USB Corporation, Cleveland, OH, USA) and sequenced both strands using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an ABI 3730 DNA Analyzer. Sequences were analyzed using the Sequencher 4.7 program (Gene Codes Corporation, Ann Arbor, MI, USA). The identified mutations were screened by sequencing in 93 unrelated individuals from the Centre d’Etude du Polymorphisme Humain (CEPH) family panel (all mutations) and 54 unrelated Japanese individuals (the two mutations observed in Japanese patients).
In silico analysis of the c.1030-9G>A mutation was performed with two exon prediction programs. GENSCAN is available at http://genes.mit.edu/GENSCAN.html and GrailEXP Exon Prediction Program (Perceval) at http://compbio.ornl.gov/grailexp/. We analyzed the c.1030-9G>A mutation from patient and control RNA extracted from fibroblast cultures with RNeasy mini kit (Qiagen, Hilden, Germany). The c.936dupG mutation was analyzed from freshly obtained peripheral blood using the PAXgene Blood RNA System (PreAnalytiX GmbH, Hombrechtikon, Switzerland). The reverse transcriptase (RT)-PCR was performed with High-Capacity cDNA Archive Kit (Applied Biosystems). We amplified the cDNA with primers located in exons 6 and 10 and sequenced the products as described above.
Construction of the expression plasmid and site-directed mutagenesis
The ORF of the human SIL1 cDNA was cloned in-frame into the aminoterminal hemagglutinin (HA) tag containing pAHC expression vector (kindly provided by T Mäkelä, University of Helsinki, Finland). Two nucleotide changes, c.1370T>C (p.Leu457Pro) and c.1372A>T (p.Lys458X), were introduced into the pAHC-SIL1 construct by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The primer sequences are available on request. All constructs were verified by sequencing the entire coding regions of the inserts.
Cell culture and transfections
COS-1 cells (American Type Culture Collection, Rockville, MD, USA) were cultivated in Dulbecco's modified Eagle's medium (Lonza Walkersville Inc., Walkersville, MD, USA) supplemented with 10% fetal calf serum (PromoCell GmbH, Heidelberg, Germany), penicillin and streptomycin, and 1 × GlutaMAX (Gibco, Invitrogen, Carlsbad, CA, USA). The cells (1.5 × 105) were plated onto six-well plates on coverslips 1 day prior to transfection, and were then transfected with 2 μg of wild-type or mutated pAHC-SIL1 using FuGENE 6 Transfection Reagent (Roche, Indianapolis, IN, USA) according to the manufacturer's instructions. For immunofluorescence analysis, protein production was inhibited by a 2 h incubation with cycloheximide 18 h after transfection, and cells were fixed with 4% paraformaldehyde.
Immunofluorescence staining, antibodies, and microscopy
Transfected COS-1 cells were permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 15 min or with 0.2% saponin in PBS supplemented with 0.5% bovine serum albumin (BSA) for 30 min. Mouse or rabbit anti-HA (Covance Research Products, USA; clone 16B12, 1:16 000; and Santa Cruz Biotechnology, Santa Cruz, CA, USA; Y-11, 1:500, respectively) antibodies were used to detect the N terminus of the HA-tagged SIL1 protein. To differentiate subcellular compartments, rabbit anti-GRP78 (Sigma-Aldrich Co., St Louis, MO, USA; 1:100) and mouse anti-PDI (Stressgen, Assay Designs Inc., Ann Arbor, MI, USA; 1:50) antibodies were used for ER, rabbit anti-p58 (kindly provided by R Pettersson, Ludwig Institute for Cancer Research, Sweden) was used for ERGIC, sheep anti-TGN46 (Serotec; 1:200) and rabbit anti-Giantin (BioSite, San Diego, CA, USA; 1:1000) were used for Golgi apparatus, and mouse anti-Lamp-1 (DSHB, H4A3; developed by August JT and Hildreth JEK and obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA, USA; 1:100) was used for lysosomes. Secondary antibodies used were Cy2- or Cy3-conjugated anti-rabbit IgG, Cy2-conjugated anti-goat IgG, and Cy2- or Cy3-conjugated anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA, USA; 1:200). The cells were examined and photographed with Axioplan 2 microscope and AxioVision 3.1 (Carl Zeiss Inc., Oberkochen, Germany).
Primary neuronal cultures and immunofluorescence analysis
Hippocampi of embryonic day 16.5 C57BL mice were dissected out in ice-cold 100 mM PBS without Ca2+/Mg2+ (PBS, pH 7.4) supplemented with 20 mM glucose. Tissue was trypsinized by adding 20 ng/ml of DNAseI and 0.1%/0.4 mM trypsin-EDTA and by incubating for 5 min at 37°C. Trypsin was inhibited with fetal calf serum and the tissue was further mechanically dissociated. Four hippocampi were pooled and resuspended in a Neurobasal medium (Gibco, BRL) supplemented with 1 × B27 (Gibco), glutamine, and antibiotics. The cells were plated on poly-D-lysine (Sigma-Aldrich), coated 58 mm dishes with coverslips, and cultured for 8–14 days. The cells were fixed in 4% paraformaldehyde in PBS for 20 min and permeabilized with 0.1% Triton X-100 in PBS containing 0.5% BSA. The primary antibodies used were goat anti-SIL1 (Abcam Ltd, Cambridge, UK; 1:5), rabbit anti-GRP78 (Sigma-Aldrich; 1:300), and rabbit anti-PDI (Stressgen; 1:50). The secondary antibodies used were Cy2-conjugated anti-goat-IgG and Cy3-conjugated anti-rabbit-IgG (Jackson ImmunoResearch; 1:250). The cells were examined and photographed with Axioplan 2 microscope and AxioVision 3.1 (Zeiss). The picture panels were produced with Adobe Photoshop (Adobe Systems Inc., San Jose, CA, USA).
Results
Mutation analysis
We identified four novel mutations in SIL1 in five families with classical MSS (Table 3). All mutations cosegregated with the disease phenotype in the respective families. In patients M2303 and M2304, we identified a homozygous one-base duplication in exon 9, c.936dupG, affecting a stretch of six G nucleotides (Figure 1a). In cDNA analysis, the mutant transcript showed normal splicing of exons 9 and 10 (data not shown). The mutation is predicted to cause frameshift at codon 313 and truncation of the protein after 38 novel amino acids in the beginning of the last exon. Patient M2003 was homozygous for a nonsense mutation c.1367T>A in exon 10, which created a premature stop codon (p.Leu456X) deleting six amino acids from the end of the protein (Figures 1b and 2). The missense mutation, c.1370T>C, was identified in two affected siblings M2103 and M2104. It changes a non-conserved leucine to a proline (p.Leu457Pro) in exon 10 (Figures 1c and 2). The Norwegian patients (M704, M1303, M1304), without either known consanguinity within the families or known relation between the two families, were homozygous for a splice-acceptor site mutation c.1030-9G>A (Figure 1d). The mutation affects a sequence, which is highly similar to the normally active 3′ splice site in intron 9, and introduces a novel splice site (Figure 1 d). In silico analysis comparing the normal and mutant sequence predicted that the novel splice site is stronger than the wild-type one. Consequently, in RT-PCR analysis, an insertion of seven nucleotides was detected in mature mRNA (Figure 1e). This insertion is predicted to cause a frameshift at codon 345 and truncation of the protein after eight novel amino acids (p.Phe345AlafsX9). We identified one carrier of the c.936dupG among 54 Japanese controls, whereas other mutations were not detected in controls.
The ER retrieval tetrapeptide of SIL1. The upper panel shows how the c.1367T>A (p.Leu456X) and c.1370T>C (p.Leu457Pro) patient mutations together with the artificial mutant deleting the tetrapeptide (p.Lys458X) alter the polypeptide. The comparison between the C-terminal amino-acid sequence of SIL1 in different species shows that the putative ER retrieval signal (highlighted in gray) has undergone many changes during evolution. The motif is in its consensus form, KDEL, only in the mosquito while different yeasts carry similar motifs, RDEL or ADEL. A star marks amino acids identical in all sequences in the alignment. The alignment was carried out by MAFFT version 6.500a.
Subcellular localization of exogenously expressed wild-type and mutant SIL1 proteins
We transiently transfected COS-1 cells with wild-type and mutant HA-tagged SIL1 constructs. The wild-type SIL1 protein repeatedly colocalized with ER markers GRP78 (Figure 3a–c) and PDI, an ERGIC marker p58 (data not shown), and Golgi apparatus markers TGN46 and Giantin (Figure 3d–f). Both the missense mutation detected in patients (p.Leu457Pro) and an artificial mutation deleting the predicted ER retrieval sequence (p.Lys458X) resulted in altered subcellular localization of mutant SIL1. Two different staining patterns, which were dependent on the protein expression level, were detected. In transfected cells with higher expression level, both mutant proteins were seen in aggregates (Figure 3g and m) that displayed strongest overlap with GRP78 and PDI markers, suggesting ER localization (Figure 3i). The aggregates did not overlap with ERGIC (Figure 3o), Golgi, or lysosome markers (data not shown). In cells with lower expression level, the mutant SIL1 was detected in the ER without aggregate formation (Figure 3j–l). Over half of both p.Leu457Pro and p.Lys458X expressing cells showed the aggregates, while the ER-staining pattern was detected in less than half of the cells.
Subcellular localization of wild-type and mutant SIL1. (a–f) Distribution of HA-tagged wild-type SIL1 in transiently transfected COS-1 cells. HA immunoreactivity is shown in red (a) and the ER-resident chaperone GRP78 immunoreactivity in green (b). Colocalization of the two proteins is shown in yellow (c). HA immunoreactivity is shown in red and is toned down by shortening the exposure time to obtain better visualization of Golgi (d). Golgi apparatus marker Giantin immunoreactivity is shown in green (e). HA-tagged wild-type SIL1 colocalizes with Giantin as indicated by the yellow color (f). (g–o) Distribution of HA-tagged mutant SIL1 in transiently transfected COS-1 cells. HA-tagged Lys458X mutant SIL1 forms aggregates (g), which show overlap with GRP78 (i). Similarly, aggregates are present in cells expressing the HA-tagged Leu457Pro mutant protein (j; the upper cell), and cells with lower expression levels show ER staining without aggregation formation (j; the lower cell). ER marker PDI is shown in green (k) and the overlay of two pictures (l) shows that the aggregates are present in the ER. The Leu457Pro mutant protein aggregates do not overlap with ERGIC marker p58 shown in green (n).
Subcellular localization of endogenous SIL1 in primary mouse hippocampal neurons
We investigated the subcellular localization of endogenous SIL1 in mouse neurons. Using immunofluorescence analysis, we found SIL1 to colocalize with PDI (Figure 4a–c) and GRP78 (data not shown) in cultured mouse primary hippocampal neurons, indicating ER localization. Endogenous SIL1 did not overlap with the Golgi marker Giantin (data not shown).
Subcellular localization of endogenous SIL1 in mouse hippocampal neurons. Immunofluorescence analysis reveals partial colocalization of SIL1 and ER marker PDI in cultured mouse hippocampal neurons. (a) PDI shows cellular staining compatible with the known ER localization. (b) SIL1 expression is indicated in green. (c) An overlay of panels a and b, where yellow indicates colocalization.
Candidate gene analysis
We sequenced from genomic DNA of 18 patient samples and one control sample the eight coding exons of the HSPA5 gene, the 26 coding exons of the HYOU1 gene, and the 20 coding exons of the AARS gene together with at least 20 base pairs of flanking intronic sequences. No disease-causing mutations were identified.
Discussion
We report four novel homozygous mutations in SIL1 in five families with classical MSS. So far, all reported MSS-associated mutations have been predicted to cause loss of SIL1 function either through nonsense and frameshift alterations or in-frame deletions of putative functionally important amino acids.5, 6, 7 Three of the mutations identified here (c.936dupG, c.1030-9G>A, and c.1367T>A) are predicted to cause truncation of the protein product within the amino acids encoded from the last SIL1 exon through a frameshift or a nonsense alteration. Because there are no exon–exon junctions downstream from the premature termination codon that could initiate nonsense-mediated decay,20 these mutations are likely to result in translation of polypeptides. Similarly, a protein product can be anticipated to be present in the cells of the patients with the fourth identified mutation, the missense substitution p.Leu457Pro.
Given that the mutant protein products are stable within the cells, the mechanism through which the Leu457Pro and Leu456X mutant proteins mediate their deleterious effect is of interest, as both affect the C-terminal region of the 461 amino acid SIL1. In the p.Leu456X mutant, the last six amino acids, including the putative ER retrieval tetrapeptide, KELR (Figure 2), are deleted. In p.Leu457Pro, the substitution of a leucine to a cyclic proline may influence the tertiary structure of SIL1 by making a turn in the polypeptide and hiding the putative ER retrieval tetrapeptide. We thus hypothesized that the Leu456X and Leu457Pro mutant proteins are likely to lead to defective retrieval of the mutant protein from later stages of the secretory pathway. In order to test this hypothesis, we performed double immunofluorescence analysis in transiently transfected COS-1 cells. We first tested the localization of wild-type SIL1. Contrary to the previous studies that have reported ER localization,8, 11 we detected SIL1 in both the ER and the Golgi apparatus. We then investigated the subcellular localization of endogenous SIL1 in mouse hippocampal neurons, where we previously showed SIL1 expression.4 Endogenous SIL1 was detected in the ER, but no colocalization was detected with a Golgi marker. Thus, the localization of wild-type overexpressed SIL1 in COS-1 cells is likely to be due to overproduction of the protein leading to the escape of the product from the ER, a phenomenon that has been seen with proteins possessing weaker ER retrieval signals.21 In SIL1, the ER retrieval motif has undergone several changes during evolution and may thus be weaker than the highly conserved motif, for example in GRP7822 or in other ER chaperones.
We then proceeded to the analysis of mutant SIL1 proteins. Since, in addition to the c.1367T>A (p.Leu456X) mutation reported here, several mutations predicting a premature stop codon in exon 10 have been described,5, 7 we created an artificial mutant deleting only the predicted ER retrieval signal at the very end of the protein product (p.Lys458X). Contrary to the hypothesis, the mutant SIL1 proteins were not detected in later stages of the secretory pathway but formed aggregates within ER in the transfected COS-1 cells. The aggregate formation was dependent on the expression level and reproducible throughout the experiments and was never present in cells overexpressing wild-type SIL1, implying that it is not an overexpression artifact. Taken together, it is likely that the two mutants are indeed stable and that their pathogenicity is explained by structural changes making them prone to aggregate formation. Interestingly, the wz mutant mouse shows ubiquitinated protein accumulations both in the ER and in the nucleus of the affected cells prior to cerebellar Purkinje cell degeneration.11 The biochemical composition of these aggregates, however, remains unknown.
We sequenced three functional candidate genes for mutations in 18 patients negative for SIL1 mutations. HSPA5 encoding GRP78 and HYOU1 were selected because of known interaction or overlapping function with SIL1.16, 23 Although both HSPA5 and HYOU1 mouse knockout models show embryonic lethality,24 mutations leading to for example reduced protein expression or altered protein function could result in MSS phenotype. The AARS gene was considered a candidate on the basis of similar cerebellar phenotype in mouse models. However, we did not find any disease-causing alterations in these genes. The possibility remains that in a subset of patients, a mutation is located in the intronic or regulatory regions that our sequencing strategy did not cover. Nevertheless, mutations in HSPA5, HYOU1, and AARS are unlikely to be major underlying causes for MSS.
The SIL1 mutation-positive patients show almost invariably the hallmark clinical features of cerebellar atrophy and ataxia, cataracts, mental retardation, and myopathy (Table 1). The additional features and their severity vary from patient to patient, and some rare findings, for example the atrophy of pons, may be present. We identified no SIL1 mutations in a subset of patients that fulfilled the clinical criteria for classical MSS. This further supports the previous suggestion of genetic heterogeneity in MSS. Moreover, many of the mutation-negative patients did not show all the cardinal features (Table 2) and some presented with additional symptoms, such as optic atrophy and epilepsy, not typically present in SIL1 mutation-positive MSS patients. Further genes need to be identified to unravel the molecular genetic basis of MSS and MSS-like disorders.
Taken together, our findings strengthen the previous findings of loss-of-function mutations in SIL1 being the major cause of MSS. Moreover, our data imply that lost SIL1 function may be mediated through aggregation of mutant SIL1. Finally, the genetic background of a subgroup of MSS patients remains uncovered.
Accession codes
References
Lagier-Tourenne C, Tranebaerg L, Chaigne D et al: Homozygosity mapping of Marinesco–Sjogren syndrome to 5q31. Eur J Hum Genet 2003; 11: 770–778.
Marinesco G, Draganesco S, Vasiliu D : Nouvelle maladie familiale caractérisée par une cataracte congénitale et un arrêt du développement somato-neuro-psychique. Encéphale 1931; 26: 97–109.
Sjögren T : Hereditary congenital spinocerebellar ataxia accompanied by congenital cataract and oligophrenia. Confin Neurol 1950; 10: 293–308.
Anttonen AK, Mahjneh I, Hamalainen RH et al: The gene disrupted in Marinesco–Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet 2005; 37: 1309–1311.
Senderek J, Krieger M, Stendel C et al: Mutations in SIL1 cause Marinesco–Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet 2005; 37: 1312–1314.
Annesi G, Aguglia U, Tarantino P et al: SIL1 and SARA2 mutations in Marinesco–Sjogren and chylomicron retention diseases. Clin Genet 2007; 71: 288–289.
Karim MA, Parsian AJ, Cleves MA et al: A novel mutation in BAP/SIL1 gene causes Marinesco–Sjogren syndrome in an extended pedigree. Clin Genet 2006; 70: 420–423.
Chung KT, Shen Y, Hendershot LM : BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem 2002; 277: 47557–47563.
Tyson JR, Stirling CJ : LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J 2000; 19: 6440–6452.
Hendershot LM : The ER function BiP is a master regulator of ER function. Mt Sinai J Med 2004; 71: 289–297.
Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL : Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet 2005; 37: 974–979.
Todorov A : [Marinesco–Sjogren syndrome. 1st anatomo-clinical study]. J Genet Hum 1965; 14: 197–233.
Mahloudji M, Amirhakimi GH, Haghighi P, Khodadoust AA : Marinesco–Sjogren syndrome. Report of an autopsy. Brain 1972; 95: 675–680.
Skre H, Berg K : Linkage studies on Marinesco–Sjogren syndrome and hypergonadotropic hypogonadism. Clin Genet 1977; 11: 57–66.
Lee JW, Beebe K, Nangle LA et al: Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 2006; 443: 50–55.
Weitzmann A, Volkmer J, Zimmermann R : The nucleotide exchange factor activity of Grp170 may explain the non-lethal phenotype of loss of Sil1 function in man and mouse. FEBS Lett 2006; 580: 5237–5240.
Zoghbi HY, Orr HT : Glutamine repeats and neurodegeneration. Annu Rev Neurosci 2000; 23: 217–247.
Andersen B : Marinesco–Sjoegren Syndrome: Spinocerebellar Ataxia, Congenital Cataract, Somatic and Mental Retardation. Dev Med Child Neurol 1965; 47: 249–257.
Torbergsen T, Aasly J, Borud O, Lindal S, Mellgren SI : Mitochondrial myopathy in Marinesco–Sjogren syndrome. J Ment Defic Res 1991; 35 (Part 2): 154–159.
Maquat LE : Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 2004; 5: 89–99.
Lewis MJ, Pelham HR : Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell 1992; 68: 353–364.
Munro S, Pelham HR : A C-terminal signal prevents secretion of luminal ER proteins. Cell 1987; 48: 899–907.
Steel GJ, Fullerton DM, Tyson JR, Stirling CJ : Coordinated activation of Hsp70 chaperones. Science 2004; 303: 98–101.
Ni M, Lee AS : ER chaperones in mammalian development and human diseases. FEBS Lett 2007; 581: 3641–3651.
Acknowledgements
We thank the patients, their families, and referring clinicians for collaboration. Hanna Olanne, Ann-Liz Träskelin, and Mira Aronen are thanked for their excellent technical assistance. Professor Michel Koenig, Dr Clotilde Lagier-Tourenne, Dr Ibrahim Mahjneh, Dr Riikka Hämäläinen, Dr Tarja Joensuu, and Dr Ulla Lahtinen are thanked for help and advice. Dr EAJ Peeters is thanked for providing clinical data on patient M2003. This study was supported by the Folkhälsan Research Foundation and the Center of Excellence in Complex Disease Genetics of the Academy of Finland for A-EL, and by the Biomedicum Helsinki Foundation, Emil Aaltonen Foundation, Finnish Medical Foundation, and Neurologiasäätiö for A-KA. A-KA and ES are fellows of the Helsinki Biomedical Graduate School.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Anttonen, AK., Siintola, E., Tranebjaerg, L. et al. Novel SIL1 mutations and exclusion of functional candidate genes in Marinesco–Sjögren syndrome. Eur J Hum Genet 16, 961–969 (2008). https://doi.org/10.1038/ejhg.2008.22
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.22
Keywords
This article is cited by
-
A Review of Brain and Pituitary Gland MRI Findings in Patients with Ataxia and Hypogonadism
The Cerebellum (2023)
-
Novel Ocular Features in a Child with Marinesco-Sjögren Syndrome: Case Report and Literature Review
SN Comprehensive Clinical Medicine (2021)
-
Ataxia in children: early recognition and clinical evaluation
Italian Journal of Pediatrics (2017)
-
Cellular Signature of SIL1 Depletion: Disease Pathogenesis due to Alterations in Protein Composition Beyond the ER Machinery
Molecular Neurobiology (2016)
-
The nucleotide exchange factor SIL1 is required for glucose-stimulated insulin secretion from mouse pancreatic beta cells in vivo
Diabetologia (2014)