The bacterium Clostridioides difficile can cause severe infection in the intestines.Credit: D. Phillips/SPL
When Melody Smith began her fellowship in haematology and oncology at the Memorial Sloan Kettering Cancer Center in New York, she had no idea that she would soon be writing her own code, working with computational biologists and analysing faecal samples to understand the microbial composition — or microbiome — of the gut.
Now at Stanford University in California, Smith is one of a growing number of researchers exploring how the complex and diverse populations of microbes in the body — known as the microbiota — influence how people respond to cancer treatments.
“Before I started my training, I wouldn’t have thought much about the microbiome,” Smith says. “But it’s been really interesting to get to know the field and to understand all of the different diseases where the microbiome is shown to be relevant, especially in the field of oncology.”
Part of Nature Career Guide: Cancer
Cancer treatment is no longer the domain just of oncologists. It now also involves specialists in microbiology, artificial intelligence, diet and nutrition, genomics, bioinformatics and computing. Their work is revealing how the gut microbiome can make the difference between treatment success or failure.
Smith’s journey into the world of the microbiome mirrors the expansion of the field generally. It began when she worked with Sloan Kettering medical oncologist Marcel van den Brink, who had long been interested in improving the outcomes of bone-marrow transplants.
Van den Brink knew of studies done in the 1970s and 1980s suggesting that the gut microbiome affected whether people who received bone-marrow transplants developed a potentially lethal condition called graft versus host disease, in which the transplanted cells mount an immune response against the recipient.
So, in 2009, he and his colleagues began a quest to understand the role of the gut microbiome in the outcomes not just of bone-marrow transplants, but also of other immune-based treatments, including checkpoint inhibitors and T-cell therapy.
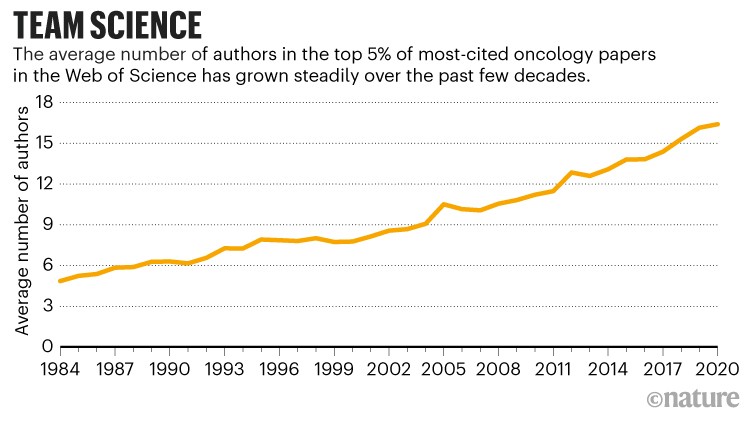
That quest has seen van den Brink expand his laboratory from just a few postdoctoral fellows, PhD students and technicians to more than 40 members from a wide range of disciplines, including 4 computational biologists — a trend that is reflected in the growing interdisciplinarity in the authorship of cancer papers (see ‘Team science tackles the microbiome’).
Numbers game
Van den Brink was keen to involve computational biologists because studying the microbiome involves analysing vast amounts of genomic data. It used to be that the composition of the gut microbiota could be determined only by growing the organisms in culture, and many species couldn’t be grown in this way. Now, researchers can tease apart the genomes of every species in a sample. But doing so requires enormous computing power.
“Sequencing in this case will give us millions of little readings of the DNA,” says computational biologist Nicola Segata in the Department of Cellular, Computational and Integrative Biology at the University of Trento in Italy. “So the computation part of the story here is to try to make sense of these millions and millions for each sample.”
One way to identify the organisms is to compare the sections of DNA with the genomes of known species to look for matches. But around 1,000 species of microbe are known to dwell in the human gut, and there are probably many more yet to be identified. A second method involves trying to piece together the DNA fragments to make whole genomes — rather like trying to solve lots of jigsaw puzzles simultaneously after mixing all the pieces together, Segata says. And today’s computing power is struggling to keep up with all the pieces in these puzzles, “so we need computational people to think about smart algorithms to solve them”.
Artificial intelligence and machine learning, for instance, are being used not only to assemble genomes, but also to understand how microbial species are interacting and affecting human health, says Leo Lahti, a data scientist at the University of Turku in Finland.
“If you look at just communities of microbial species, that’s one aspect,” he says. But studies of the microbiome are now moving beyond simply listing what species are present, to exploring their functions and interactions, with each other and with their host, so the field is becoming even more complex, Lahti says.
AI finds microbial signatures in tumours and blood across cancer types
Many machine-learning approaches can be used to interpret these vast amounts of data. The challenge for researchers is knowing which ones to use. “More important than any particular tool is to have the skill to understand the basic principles of different tools and techniques, and the ability to combine them into reproducible workflows in a new ways,” Lahti says. The challenge is so big that it takes more than one researcher or even one team; data scientists are now working together in an open, collaborative way, so that everyone can make use of what has been developed and learnt by others.
“For me personally, one of the most exciting aspects of this has been the ability to join the open data-science communities, and be a part of this development and benchmarking and evaluation of all these different machine-learning and statistical techniques,” Lahti says. “It’s not about working alone on a single tool, but being part of a community.”
All this means that the field is crying out for people with some background in computational science, Segata says. “There is a lack of these skill sets, because I think that people that are putting themselves in computer science, they probably have other things in mind,” he says. “We should try to attract computer scientists more, to tell them they can have a huge impact in life sciences.” Ultimately, Segata says, their programming skills can have a direct impact on public health.
Lahti says that such skills don’t necessarily need to come from training in mathematics or statistics — they can come from other fields that use computation techniques, such as physics, ecology and even economics. “You need this kind of applied angle, and then you need the robust set of different techniques.”
What you eat
Another type of expertise that is becoming important in exploring the intersection between the gut microbiome and cancer outcomes is nutrition. Jennifer Wargo, a surgical oncologist and translational scientist at the MD Anderson Cancer Center in Houston, Texas, has been looking into how diet influences the gut microbiome and cancer treatment outcomes. She and her colleagues have found that people with melanoma respond significantly better to immunotherapy if they consume a high-fibre diet than if they eat a low-fibre diet1.
She and her colleagues are now running trials in people receiving immunotherapy to find out whether diets such as high-fibre, ketogenic or intermittent fasting might increase the chances of a good response to treatment. But she’s keen for more researchers to examine this, both in clinical trials and in preclinical studies. “Are there supplements that we could derive that could actually enhance immunity and immunotherapy response, and maybe even vaccine response and promote overall health?”
Microbial culture from the gut of a potential donor for a faecal-microbiota transplant.Credit: Lewis Houghton/SPL
Van den Brink says that diet has been a surprisingly under-served area of study in cancer medicine. When doctors take people through expensive bone-marrow transplants, “we basically treat them like intensive-care patients: I know every blood count; every vital sign; and every drug and when it was given”, he says. “When it comes to diet, in many cases, I’ve scribbled ‘took half a sandwich’.”
Now in the process of setting up her own lab, Smith says her work on the gut microbiome means she will soon looking to bring together a wide range of skill sets. “I definitely want people who are going to be able to work at the bench and do experiments in the animal models, and in vitro setting, but also people who have computational skills,” she says. She’s not yet at the stage of recruiting a dedicated computational biologist, but notes that those skills are needed to analyse genome-sequencing data, “so I’ve been trying to, in my postings, look for people with that combined experience and background”.
The work has even motivated her to explore coding herself, to help her understand how the data can be analysed. “I think a lot of medical trainees are starting to realize there’s a lot of benefit to developing computational skills very early,” she says.
Faecal fascination
With greater understanding of the influence of the gut microbiome on treatment outcomes comes interest in how to tweak the microbiota to improve those outcomes. Faecal microbiota transfer — transplantation of faecal microbes from a healthy donor — can have significant benefits in treating gut conditions such as chronic infection with the bacterium Clostridioides difficile, which can cause severe diarrhoea.
“I do think there could be a potential place for faecal microbiota transplant in patients receiving cell therapy who don’t respond to it,” Smith says. Another approach, she adds, is to profile the microbiome before the therapy is infused, to see whether there are ways to boost the numbers of beneficial bacteria. She’s also interested in whether administration of just the metabolites gut bacteria produce — including butyrate and other short-chain fatty acids — could improve patient outcomes after immunotherapy.
The biotechnology sector is interested in developing more targeted approaches to altering the gut microbiome, too. And that opens up opportunities in industry — and funding — for researchers with the skills needed to characterize, understand and alter the microbiome. For example, van den Brink is involved in a clinical trial treating people with cancer with a pill containing a mixture of bacterial species known to have health benefits such as reducing inflammation. He says the food industry is also showing interest in the microbiome.
Given the individual nature of the microbiome, Wargo says that interventions to alter it and improve outcomes will probably need to take a personalized approach. “Some people will have a stellar microbiome that doesn’t need a lot and we just need to feed it the right things,” she says. “But others will, potentially — especially patients with cancer — have a disrupted gut microbiome, and they’ll either need faecal transplant or some kind of consortia to actually be able to get them back on the right track.”
Although awareness of the influence of the gut microbiome on cancer treatment outcomes has been around for decades, Wargo says these are relatively early — and exciting — days for the research field. “We’ve only scratched the surface of this,” she says. “I think there are tremendous careers to be made, and tremendous discoveries and advances in promoting overall health and the role of the microbiome.”

 Nine ‘brain food’ tips for researchers
Nine ‘brain food’ tips for researchers
 AI finds microbial signatures in tumours and blood across cancer types
AI finds microbial signatures in tumours and blood across cancer types
 Gut feeling: building a picture of Latin American microbiomes
Gut feeling: building a picture of Latin American microbiomes