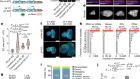
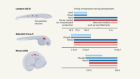
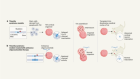
- NEWS FEATURE
These cellular clocks help explain why elephants are bigger than mice

Illustration by Alberto Seveso
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 592, 682-684 (2021)
doi: https://doi.org/10.1038/d41586-021-01086-9
References
Cooke, J. & Zeeman, E. C. J. Theor. Biol. 58, 455–476 (1976).
Palmeirim, I., Henrique, D., Ish-Horowicz, D. & Pourquié, O. Cell 91, 639–648 (1997).
Diaz-Cuadros, M. et al. Nature 580, 113–118 (2020).
Matsuda, M. et al. Nature 580, 124–129 (2020).
Chu, L.-F. et al. Cell Rep. 28, 2247–2255 (2019).
Matsumiya, M., Tomita, T., Yoshioka-Kobayashi, K., Isomura, A. & Kageyama, R. Development 145, dev156836 (2018).
Matsuda, M. et al. Science 369, 1450–1455 (2020).
Rayon, T. et al. Science 369, eaba7667 (2020).
Swovick, K. et al. Mol. Cell Proteom. 20, 100041 (2021).

 CRISPR’s hopeful monsters: gene-editing storms evo-devo labs
CRISPR’s hopeful monsters: gene-editing storms evo-devo labs
 The trickiest family tree in biology
The trickiest family tree in biology
 The labs growing human embryos for longer than ever before
The labs growing human embryos for longer than ever before