- RESEARCH HIGHLIGHT
CRISPR zeroes in on antibiotic-resistance genes
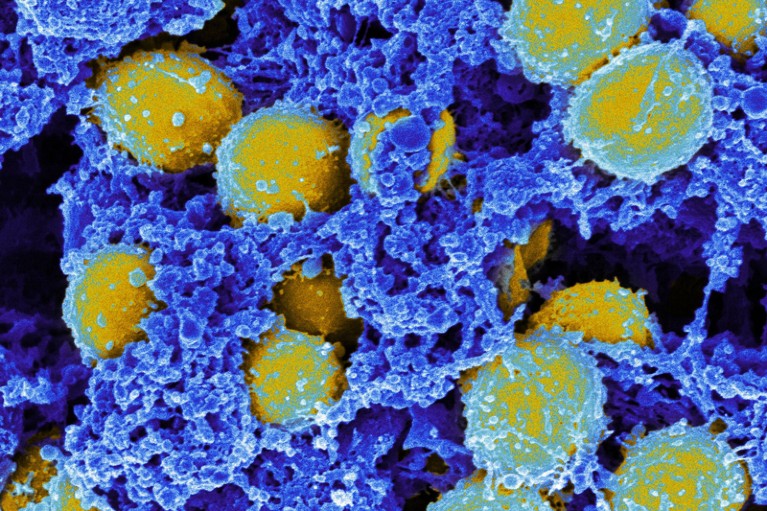
When applied to respiratory fluid, a CRISPR-based method identified a gene that confers antibiotic resistance on the bacterium Staphylococcus aureus (yellow). Credit: Cdc/Phanie/Shutterstock
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 569, 602 (2019)
doi: https://doi.org/10.1038/d41586-019-01577-w



