- NEWS AND VIEWS
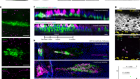
More than one way to induce a neuron
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 557, 316-317 (2018)
doi: https://doi.org/10.1038/d41586-018-04978-5
References
Zeng, H. & Sanes, J. R. Nature Rev. Neurosci. 18, 530–546 (2017).
Tsunemoto, R. et al. Nature 557, 375–380 (2018).
Ramón y Cajal, S. Histologie du Système Nerveux de l’Homme et des Vertébrés (Maloine, 1909).
Lodato, S. & Arlotta, P. Annu. Rev. Cell. Dev. Biol. 31, 699–720 (2015).
Vierbuchen, T. et al. Nature 463, 1035–1041 (2010).
Blanchard, J. W. et al. Nature Neurosci. 18, 25–35 (2015).
Treutlein, B. et al. Nature 534, 391–395 (2016).
Mi, D. et al. Science 360, 81–85 (2018).
Mayer, C. et al. Nature 555, 457–462 (2018).

 Read the paper: Diverse reprogramming codes for neuronal identity
Read the paper: Diverse reprogramming codes for neuronal identity
 A compass for stem-cell differentiation
A compass for stem-cell differentiation
 Brain versus brawn
Brain versus brawn