- NEWS AND VIEWS
Ultrasound approach tracks gut microbes
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 553, 36-37 (2018)
doi: https://doi.org/10.1038/d41586-017-08911-0
References
Bourdeau, R. W. et al. Nature 553, 86–90 (2018).
Walter, J. & Ley, R. Annu. Rev. Microbiol. 65, 411–429 (2011).
Berlec, A., Završnik, J., Butinar, M., Turk, B. & Štrukelj, B. Microb. Cell Fact. 14, 181 (2015).
Hong, G., Antaris, A. L. & Dai, H. Nature Biomed. Eng. 1, 0010 (2017).
Walsby, A. E. Microbiol. Rev. 58, 94–144 (1994).
Shapiro, M. G. et al. Nature Nanotechnol. 9, 311–316 (2014).
Sonnenborn, U. & Schulze, J. Microb. Ecol. Health Dis. 21, 122–158 (2009).
Din, M. O. et al. Nature 536, 81–85 (2016).
Danino, T., Lo, J., Prindle, A., Hasty, J. & Bhatia, S. N. ACS Synth. Biol. 1, 465–470 (2012).
Weissleder, R. Nature Rev. Cancer 2, 11–18 (2002).
Weber, W. & Fussenegger, M. Nature Rev. Genet. 13, 21–35 (2012).
Xu, M. & Wang, L. V. Rev. Sci. Instrum. 77, 041101 (2006).
Brenner, K., You, L. & Arnold, F. H. Trends Biotechnol. 26, 483–489 (2008).
Macía, J., Posas, F. & Solé, R. V. Trends Biotechnol. 30, 342–349 (2012).
Ettema, C. H. & Wardle, D. A. Trends Ecol. Evol. 17, 177–183 (2002).

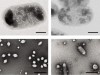 Read the paper: Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts
Read the paper: Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts
 An intracellular dance visualized
An intracellular dance visualized
 Bacteria synchronized for drug delivery
Bacteria synchronized for drug delivery