Abstract
Lung squamous cell carcinoma (SCC) is one of the major subtypes of lung cancer. Our current knowledge of oncogenic drivers in this specific subtype of lung cancer is largely limited compared with lung adenocarcinoma (ADC). Through exon array analyses, molecular analyses and functional studies, we here identify the TRA2B-DNAH5 fusion as a novel oncogenic driver in lung SCC. We found that this gene fusion occurs exclusively in lung SCC (3.1%, 5/163), but not in lung ADC (0/119). Through mechanistic studies, we further revealed that this TRA2B-DNAH5 fusion promotes lung SCC malignant progression through regulating a SIRT6-ERK1/2-MMP1 signaling axis. We show that inhibition of ERK1/2 activation using selumetinib efficiently inhibits the growth of lung SCC with TRA2B-DNAH5 fusion expression. These findings improve our current knowledge of oncogenic drivers in lung SCC and provide a potential therapeutic strategy for lung SCC patients with TRA2B-DNAH5 fusion.
Similar content being viewed by others
Introduction
Lung cancer, which can be divided into small cell lung cancer (SCLC) and non-SCLC (NSCLC) according to the histology differences, is the leading cause of cancer-related death worldwide1. NSCLC accounts for about 85% of all lung cancer cases, with a 5-year survival rate as low as 16%, and is composed of two major subtypes including adenocarcinoma (ADC) and squamous cell carcinoma (SCC).
Currently, although the traditional therapeutic methods including surgery, chemotherapy and radiotherapy are widely used for lung cancer patients, the effective treatment options are still limited. Molecular targeted therapy, which has better effect and less side effect, is a more promising therapeutic method for lung cancer patients as compared with traditional methods. The identification of oncogenic drivers such as EGFR mutation has significantly reformed the current strategies for NSCLC treatment in clinic and initiated the era of molecular targeted therapy2. Searching for new oncogenic drivers in NSCLC also becomes increasingly important for the development of molecular targeted therapy.
To date, many genetic alterations, such as KRAS mutation3, EGFR mutation4,5,6, PDGFRA amplification7 and DDR2 mutation8, have been identified as oncogenic drivers in NSCLC. EGFR is an essential therapeutic target which is fully studied, and patients with EGFR mutations can benefit from EGFR inhibitors4,5,6. Fusion genes, including ALK, ROS1, RET, NTRK1, NRG1 and FGFR fusions, have been identified to be a new class of oncogenic drivers in NSCLC in recent years9,10,11,12,13,14,15,16,17,18. ALK fusion, ROS1 fusion and RET fusion have been proved to be essential therapeutic targets in NSCLC12,14,15,19,20,21,22. Thus, searching for new oncogenic gene fusions is also a direction for us to identify novel therapeutic targets in NSCLC.
Interestingly, oncogenic drivers have been well studied in lung ADC23,24,25,26, whereas in lung SCC, the current knowledge of oncogenic drivers is largely limited. Up to now, only a few oncogenic drivers such as PDGFRA amplification, DDR2 mutation and FGFR fusions have been identified in lung SCC7,8,17, which significantly limits the molecular targeted therapy for lung SCC patients.
Using previously established method for gene fusion detection based on exon array analyses23,27, as well as molecular analyses and functional studies, we here identify the TRA2B-DNAH5 fusion as a new oncogenic driver in human lung SCC. We further show that the TRA2B-DNAH5 fusion promotes lung SCC malignant progression through modulating SIRT6, activating ERK1/2 and upregulating MMP1 expression. This study indicates that the MEK/ERK cascade might serve as a potential therapeutic target for the lung SCC patients harboring TRA2B-DNAH5 fusion.
Results
Identification of TRA2B-DNAH5 fusion by exon array analyses in human lung SCC
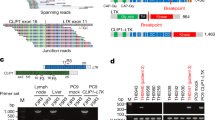
To search for novel gene fusions, as previously described23,27, we performed exon array (Affymetrix Exon 1.0) analyses in 143 NSCLC specimens (78 ADC and 65 SCC) plus 15 paired pathologically normal lung tissues. We initially identified 693 candidates (409 in ADC and 284 in SCC) as potential gene fusions or isoforms (Supplementary information, Tables S1 and S2). We then narrowed down the candidates through the following 3 steps: (1) manually go through the heatmaps and choose these candidates with obvious breakpoints; (2) exclude those candidates also detected in normal lung samples; (3) exclude those known gene isoforms by referring to the NCBI database. Eventually, we obtained 15 candidates which were potentially new gene fusions or isoforms (Table 1). We then performed 5′ RACE or 3′ RACE followed by Sanger sequencing to confirm these potential candidates, and found that most of these were novel isoforms, and only three were gene fusions (Table 1). Besides the CCDC6-RET fusion that we previously reported23, we found another two in-frame novel gene fusions, PTPRF-TIE1 fusion in lung ADC and TRA2B-DNAH5 fusion in lung SCC (Table 1, Figure 1 and Supplementary information, Figure S1).
Identification of TRA2B-DNAH5 fusion by exon array analyses in lung SCC. (A) Exon array analyses of 65 lung SCC specimens and 5 paired normal lung tissues identified a potential DNAH5 fusion in lung SCC sample 207LC. The potential fusion site was indicated by the arrow. (B) 5′ RACE analyses of 207LC showed a PCR band about 750 bp, which is obviously different from the wild-type DNAH5 band (about 500 bp). (C) Sequencing result confirmed the TRA2B-DNAH5 fusion in lung SCC sample 207LC. (D) The schematic diagram of TRA2B-DNAH5 fusion protein. (E) Detection of TRA2B-DNAH5 fusion in 163 human lung SCC specimens through RT-PCR analyses. Besides 207LC, another 4 human lung SCC specimens harbor the TRA2B-DNAH5 fusion. ***P < 0.001. LC, lung cancer; N, normal lung tissue.
We first analyzed PTPRF-TIE1 fusion. TIE1 is a transmembrane receptor tyrosine kinase, which may modulate the activity of angiopoitin-TIE2 signaling pathway and contribute to the regulation of angiogenesis28. We found that its C-terminal portion including the whole kinase domain was fused to PTPRF N-terminal part to form a new fusion protein in one “pan-negative” lung ADC specimen (Supplementary information, Figure S1). We tried to detect PTPRF-TIE1 fusion in another 47 “pan-negative” lung ADC specimens through RT-PCR analyses, but found no more samples positive for this fusion (data not shown). Cell transformation assay also showed that the PTPRF-TIE1 fusion cannot transform Ink4a−/− MEFs (data not shown), suggesting that this fusion might not be oncogenic.
We next analyzed TRA2B-DNAH5 fusion. DNAH5 is one member of axonemal dyneins, which causes sliding of microtubules in the axonemes of cilia and flagella and is found only in cells that have these two structures. We found that its C-terminal portion was fused to TRA2B N-terminal part to form a new fusion protein (Figure 1A-1D). Through long-range PCR, we identified the breakpoint at genomic DNA level (Supplementary information, Figure S2). Through RT-PCR analyses followed by Sanger sequencing, we identified another 4 specimens positive for the TRA2B-DNAH5 fusion in another cohort of 98 lung SCC specimens (Figure 1E and Supplementary information, Figure S3). Moreover, this fusion is a somatic event which does not exist in paired normal lung tissue (Figure 1E). In total, 3.1% (5/163) lung SCC patients harbor the TRA2B-DNAH5 fusion. All these five patients are male and smokers (Supplementary information, Table S3). Interestingly, no TRA2B-DNAH5 fusion was detected in 119 lung ADC through RT-PCR analyses (data now shown), indicating the exclusive existence of TRA2B-DNAH5 fusion in human lung SCC.
TRA2B-DNAH5 fusion is a novel oncogenic driver in lung SCC
We then functionally characterized the TRA2B-DNAH5 fusion in lung SCC. First, TRA2B-DNAH5 fusion significantly increased the number of foci which possess the transformation phenotype in Ink4a−/− MEFs (Supplementary information, Figure S4). Thus we proved that the TRA2B-DNAH5 fusion was capable of promoting cell transformation. Consistently, TRA2B-DNAH5 fusion promoted anchorage-independent growth of human bronchial epithelial cells (Beas2B) and human lung SCC cells (CRL-5889; Figure 2A-2D). Moreover, TRA2B-DNAH5 fusion promoted matrigel invasion of Beas2B and CRL-5889 cells (Figure 2E and 2F). TRA2B-DNAH5 fusion also significantly promoted tumor growth in CRL-5889 xenograft assay (Figure 3A-3C). A higher cell proliferation rate was observed in CRL-5889 xenograft tumors with TRA2B-DNAH5 fusion expression (Supplementary information, Figure S5A). Since Beas2B cells were not tumorigenic in immuno-deficient mice, we used another lung SCC cell line HTB-58 which has weak tumorigenic capability in xenograft assay. Consistently, the TRA2B-DNAH5 fusion could significantly promote tumor growth in HTB-58 xenograft assay (Figure 3D-3F). These results together indicate that the TRA2B-DNAH5 fusion is a novel oncogenic driver in lung SCC.
TRA2B-DNAH5 fusion promotes lung SCC progression in vitro. (A) Western blot analysis confirmed the expression of TRA2B-DNAH5 fusion in Beas2B cells. Myc tag was used to mark the TRA2B-DNAH5 fusion. (B) The TRA2B-DNAH5 fusion promoted anchorage-independent growth of Beas2B cells in soft agar assay. Statistical analysis of colony formation was shown on the right. (C) Western blot analysis confirmed the expression of TRA2B-DNAH5 fusion in CRL-5889 cells. (D) The TRA2B-DNAH5 fusion promoted anchorage-independent growth of CRL-5889 cells in soft agar assay. Statistical analysis of colony formation was shown on the right. (E, F) TRA2B-DNAH5 fusion promoted cell invasion of Beas2B cells (E) and CRL-5889 cells (F) in transwell assay. Statistics analyses were shown on the right. All data were shown as mean ± SEM. **P < 0.01 and ***P < 0.001. Ctrl, control; TD, TRA2B-DNAH5 fusion.
TRA2B-DNAH5 fusion promotes lung SCC progression in vivo. (A-C) TRA2B-DNAH5 fusion promoted tumor growth in CRL-5889 xenograft assay. Photos of xenograft tumors (A), tumor volume (B) and tumor weight (C) were shown. Ctrl, n = 6; TD, n = 6. (D-F) TRA2B-DNAH5 fusion promoted tumor growth in HTB-58 xenograft assay. Photos of xenograft tumors (D), tumor volume (E) and tumor weight (F) were shown. Ctrl, n = 7; TD, n = 7. All data were shown as mean ± SEM. **P < 0.01 and ***P < 0.001. Ctrl, control; TD, TRA2B-DNAH5 fusion.
TRA2B-DNAH5 fusion promotes lung SCC progression through upregulating MMP1 expression
To explore the potential mechanism, we comparatively analyzed the gene expression profiles of xenograft tumors with or without TRA2B-DNAH5 fusion. Functional enrichment analyses of the differentially expressed genes identified several cancer-related KEGG pathways (Figure 4A). Among those genes in the most significantly enriched pathway (hsa05200: pathways in cancer), MMP1 was the top one (Figure 4B). Principal component analysis (PCA) also indicated MMP1 as one of the most important genes (Figure 4C). MMP1 is previously reported to promote tumor progression in many cancers including lung cancer29. Through real-time PCR, western blot and immunostaining analyses, we confirmed the upregulation of MMP1 by TRA2B-DNAH5 fusion in CRL-5889 cells as well as xenograft tumors (Figure 4D and 4E, Supplementary information, Figure S5B). We also obtained similar results in Beas2B cells, HTB-58 cells and HTB-58 xenograft tumors (Supplementary information, Figure S6). These results suggest that MMP1 is a potential downstream effector which may mediate the tumor-promotive function of TRA2B-DNAH5 fusion in lung SCC. To prove this, we used shRNAs to target MMP1, and found that MMP1 knockdown significantly inhibited the function of TRA2B-DNAH5 fusion in promoting cell invasion in both Beas2B cells and CRL-5889 cells (Supplementary information, Figure S7). Moreover, MMP1 knockdown dramatically inhibited the tumor-promotive function of TRA2B-DNAH5 fusion in xenograft assay (Figure 5A-5C and Supplementary information, Figure S8). Together, these data demonstrate that TRA2B-DNAH5 fusion promotes lung SCC progression potentially through upregulating MMP1 expression.
TRA2B-DNAH5 fusion significantly upregulates MMP1 expression. (A) KEGG pathway enrichment analyses. Negative log transformed (base 10) P values was visualized in the bar plot. (B) Heatmap of differentially expressed genes in the top significant KEGG pathway (hsa05200: pathways in cancer). (C) Contributions of differential genes to the 1st and 2nd principal components (PCs), as reflected by the absolute coordinates on the two axes, respectively. The point size of each gene was correlated with its contribution to PC1. (D, E) Real-time PCR quantification and western blot analysis of MMP1 expression in CRL-5889 cells (D) and xenograft tumors (E) with or without TRA2B-DNAH5 fusion expression.
Upregulated MMP1 expression mediates the tumor-promotive function of TRA2B-DNAH5 fusion. (A) Real-time PCR quantification confirmed the knockdown efficiency of MMP1 in CRL-5889 cells with TRA2B-DNAH5 fusion expression. MMP1 knockdown inhibited the tumor-promotive function of TRA2-DNAH5 fusion in CRL-5889 xenograft assay. Photos of xenograft tumors were shown. (B, C) Tumor volume (B) and tumor weight (C) were shown. Ctrl and TD, n = 5; TD + shMMP1-1, -2, -3, n = 6. All data were shown as mean ± SEM. ***P < 0.001. Ctrl, control; TD, TRA2B-DNAH5 fusion.
TRA2B-DNAH5 fusion upregulates MMP1 expression through activating ERK1/2
We next determined how TRA2B-DNAH5 fusion upregulated MMP1 expression. To this end, we initially screened six reporters including CRE, serum response element (SRE), NF-κB, glucocorticoid response element (GRE), heat-shock element (HSE), and AP1 for their responses to TRA2B-DNAH5 fusion. Our data showed that the TRA2B-DNAH5 fusion could activate three reporters including CRE, SRE and GRE reporters (Figure 6A). Interestingly, the related genes of these three reporters, including PKA, PKC, CaMKII, JNK, P38 and the MEK-ERK cascade, were known to regulate MMP1 expression30,31,32,33,34. Moreover, the JAK-STAT pathway was reported to upregulate MMP1 expression35. Therefore, we used inhibitors toward these molecules to do further screening. The inhibitors of PKA, PKC, CaMKII, JAK-STAT, MEK-ERK cascade and JNK significantly inhibited the upregulation of MMP1 expression by TRA2B-DNAH5 fusion (Figure 6B). To narrow down the potential candidates, we further performed western blot analyses to examine the activation of these molecules. The level of p-ERK1/2 was consistently and significantly upregulated by TRA2B-DNAH5 fusion in CRL-5889 cells and xenograft tumors (Figure 6C and 6D). This was further confirmed by immunostaining of xenograft tumors with TRA2B-DNAH5 fusion expression (Supplementary information, Figure S5B). Moreover, the p-ERK1/2 level was significantly upregulated by the TRA2B-DNAH5 fusion in HTB-58 cells and xenograft tumors (Supplementary information, Figure S6D and S6F). The five MEK inhibitors decreased ERK1/2 phosphorylation and most inhibitors significantly inhibited MMP1 expression (Figure 6E and 6F), further supporting that MMP1 expression was controlled by the MEK/ERK cascade. These data collectively showed that the TRA2B-DNAH5 fusion upregulated MMP1 expression through activating ERK1/2. Consistent with this, those human lung SCC specimens with TRA2B-DNAH5 fusion harbored high levels of p-ERK1/2 and MMP1 (Supplementary information, Figure S9).
TRA2B-DNAH5 fusion upregulates MMP1 expression through activating ERK1/2. (A) The TRA2B-DNAH5 fusion specifically upregulated the activity of SRE, CRE and GRE but not other reporters as indicated in HEK-293T cells. (B) Screening of kinase inhibitors including H-7 (PKA, PKG, MLCK, and PKC inhibitor), H-8 (PKA, PKG inhibitor), H-89 (PKA inhibitor), PCC (Palmitoyl-DL-carnitine Cl, PKC inhibitor), STS (Staurosporine, PKC inhibitor), KN-62 (CaMKII inhibitor), KN-93 (CaMKII inhibitor), ML-7 (MLCK inhibitor), AT9283 (Aurora Kinase inhibitor), AG-490 (JAK-2 inhibitor), ZM 449829 (JAK-3 inhibitor), CTS (Cryptotanshinone, STAT3 inhibitor), Stattic (STAT3 inhibitor), SL327 (MEK inhibitor), PD98059 (MEK inhibitor), SP600125 (JNK inhibitor), SB 202190 (p38 MAPK inhibitor), VX-702 (p38 MAPK inhibitor). P values were calculated through comparison with the untreated TD group. (C, D) Western blot analyses of p-ERK1/2, total ERK1/2, p-CaMKIIα, p-PKAα/β, p-PKCα/β, p-PKCδ/θ, p-STAT1, p-STAT2 and p-JNK in CRL-5889 cells (C) and CRL-5889 xenograft tumors (D). (E, F) Five MEK inhibitors inhibited MMP1 expression in CRL-5889 cells with TRA2B-DNAH5 fusion expression. Real-time PCR quantification (E) and western blot analysis (F) of MMP1 were performed. All data were shown as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001. Ctrl, control; TD, TRA2B-DNAH5 fusion.
TRA2B-DNAH5 fusion activates ERK1/2 through modulating SIRT6 nuclear activity
To explore how the TRA2B-DNAH5 fusion promoted ERK1/2 activation, we performed immunoprecipitation and mass spectrometry analyses (IP-MS) in CRL-5889 cells with or without TRA2B-DNAH5 fusion expression. Through this analysis, we identified 38 protein candidates which potentially interacted with the TRA2B-DNAH5 fusion protein (Supplementary information, Table S4). We then went through these 38 proteins manually and selected 15 candidates (FAM101A, FMN2, HNRNPF, SYNE1, RPA1, ZNF451, SIRT6, APOC3, TCEB1, ANXA2, PKM2, C1QA, S100A10, ACTBL2 and BEND3) which are known to regulate tumor progression for further validation. Through gene knockdown validation, we found that SIRT6 knockdown significantly increased both p-ERK1/2 and MMP1 expression levels (Figure 7A and 7B). We confirmed the interaction between TRA2B-DNAH5 fusion protein and SIRT6 through Co-immunoprecipitation (Co-IP) assay (Figure 7C). SIRT6, which functions as a histone deacetylase in nuclei, has been reported to be a tumor suppressor through inhibiting many important cancer-promotive pathways36,37,38. SIRT6 knockdown could activate ERK1/2, consistent with previous studies37,39. Through immunofluorescence, we showed that TRA2B-DNAH5 fusion protein is preferentially located in cytoplasm (Supplementary information, Figure S10). Moreover, TRA2B-DNAH5 overexpression decreased the nuclear localization of SIRT6 (Figure 7D). These data demonstrate that TRA2B-DNAH5 fusion protein may control ERK1/2 activation through modulating nuclear localization of SIRT6.
TRA2B-DNAH5 fusion activates ERK1/2 through modulating SIRT6. (A) Real-time PCR quantification confirmed the knockdown efficiency of SIRT6 in CRL-5889 cells. (B) Western blot analysis confirmed the ERK1/2 activation and MMP1 upregulation after SIRT6 knockdown. (C) Co-IP assay confirmed the interaction between TRA2B-DNAH5 fusion protein and SIRT6. (D) TRA2B-DNAH5 fusion expression decreased the nuclear localization of SIRT6. (E) TIAM1, SUV420H1 and RIN1 were upregulated in CRL-5889 xenograft tumors with TRA2B-DNAH5 fusion expression according to the microarray data. (F) Real-time PCR quantification confirmed the upregulation of TIAM1, SUV420H1, RIN1 and several IGF signaling-related genes in CRL-5889 xenograft tumors with TRA2B-DNAH5 fusion expression. (G) Real-time PCR quantification confirmed the upregulation of TIAM1, SUV420H1 and IGF2R by TRA2B-DNAH5 fusion in CRL-5889 cells. (H) Real-time PCR quantification confirmed the upregulation of TIAM1, SUV420H1, RIN1 and several IGF signaling-related genes in CRL-5889 cells after SIRT6 knockdown. All data were shown as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001. Ctrl, control; TD, TRA2B-DNAH5 fusion.
SIRT6 inhibition activates ERK1/2 through controlling the transcriptional activities of TIAM1, SUV420H1, RIN1 and IGF signaling-related genes
We next explored how SIRT6 inhibited ERK1/2 activation. From SIRT6 ChIP-Seq data set, we annotated 3 784 genes potentially targeted by SIRT6, among which 99 genes are further selected based on its upregulation by TRA2B-DNAH5 fusion expression (data not shown). Through literature searching, we found that only three candidates, including TIAM1, SUV420H1 and RIN1, have been implicated in ERK1/2 activation40,41,42,43,44. ChIP-Seq data showed that SIRT6 binds to the promoter region of TIAM1, SUV420H1 and RIN1 genes (Supplementary information, Figure S11). Our microarray data showed that TIAM1, SUV420H1 and RIN1 gene expression were upregulated by the TRA2B-DNAH5 fusion in CRL-5889 xenograft tumors (Figure 7E). This was confirmed by real-time PCR analyses (Figure 7F). Moreover, TIAM1 and SUV420H1 were significantly upregulated by TRA2B-DNAH5 fusion in CRL-5889 cells (Figure 7G). Consistently, SIRT6 knockdown upregulated TIAM1, SUV420H1 and RIN1 in CRL-5889 cells (Figure 7H). Previous data also showed that the binding of SIRT6 to the promoters of many IGF signaling-related genes inhibits their transcription, and loss of SIRT6 activates ERK1/2 through the IGF signaling pathway39. The upregulation of multiple IGF signaling-related genes was also obvious in CRL-5889 cells and xenograft tumors (Figure 7F and 7G). Consistently, SIRT6 knockdown upregulated several IGF signaling-related genes in CRL-5889 cells (Figure 7H). Among these IGF signaling-related genes which may be involved in this process, we are more interested in IGF1R and IGF2R. Through western blot analyses, we found that IGF1R instead of IGF2R was upregulated by TRA2B-DNAH5 fusion (Supplementary information, Figure S12A-S12C). Ectopic expression of TRA2B-DNAH5 fusion promoted the increase of pIGF1R and pMEK1/2 levels (Supplementary information, Figure S12C). Moreover, treatment of the IGF1R inhibitor AG-1024 can significantly inhibit pIGF1R, pMEK as well as MMP1 levels (Supplementary information, Figure S12D). These data together suggest that TIAM1, SUV420H1, RIN1 and IGF signaling-related genes may be the potential mediators between SIRT6 inhibition and ERK1/2 activation.
Targeting ERK1/2 activation using selumetinib efficiently inhibits the growth of lung SCC with TRA2B-DNAH5 fusion expression
TRA2B-DNAH5 fusion promoted lung SCC progression through activating ERK1/2 (Figure 6 and Supplementary information, Figures S5B, S6D and S6E), and human lung SCC specimens with TRA2B-DNAH5 fusion showed high level of p-ERK1/2 (Supplementary information, Figure S9). This suggests that targeting ERK1/2 activation might be a good therapeutic strategy for the lung SCC patients harboring the TRA2B-DNAH5 fusion. Among five MEK inhibitors we used in this study, selumetinib showed the strongest inhibitory effect (Figure 6E and 6F). Interestingly, the TRA2B-DNAH5 fusion expression sensitized CRL-5889 and HTB-58 cells to selumetinib treatment (Supplementary information, Figure S13). We then performed selumetinib treatments in xenograft assay. The selumetinib treatment efficiently inhibited the growth of CRL-5889 xenograft tumors expressing the TRA2B-DNAH5 fusion (Figure 8A-8C). Selumetinib treatment significantly decreased the proliferation rate of xenograft tumors (Supplementary information, Figure S14A); similarly, p-ERK1/2 and MMP1 levels were also downregulated by selumetinib treatment (Supplementary information, Figure S14B-S14D). This was further confirmed in HTB-58 xenograft assay (Figure 8D-8F). No significant weight loss was detected in mice treated with selumetinib (Supplementary information, Figure S15). Collectively, our data suggest that selumetinib might serve as a potential candidate drug for lung SCC patients harboring the TRA2B-DNAH5 fusion.
Targeting ERK1/2 activation using selumetinib efficiently inhibits the growth of lung SCC with TRA2B-DNAH5 fusion expression. (A-C) Selumetinib significantly inhibited tumor growth in CRL-5889 xenograft tumors with TRA2B-DNAH5 fusion expression. Photos of xenograft tumors (A), tumor volume (B) and tumor weight (C) were shown. The treatment lasted 15 days. Vehicle, n = 10; selumetinib, n = 11. (D-F) Selumetinib significantly inhibited tumor growth of HTB-58 xenograft tumors with TRA2B-DNAH5 fusion expression. The photos of xenograft tumors (D), tumor volume (E) and tumor weight (F) were shown. The treatment lasted 9 days. Vehicle, n = 14; selumetinib, n = 12. All data were shown as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001.
Discussion
Despite advanced knowledge of oncogenic drivers in human lung ADC, the potential oncogenic drivers in human lung SCC remain largely unknown. DDR2 gain-of-function mutation and FGFR fusions have been identified in 3.8% and 3.5% lung SCC, respectively8,45. Our study here identifies a novel oncogenic driver, the TRA2B-DNAH5 fusion, exclusively in human lung SCC but not ADC. About 3.1% lung SCC harbor the TRA2B-DNAH5 fusion, which is comparable to DDR2 mutation and FGFR fusions. Through mechanistic studies, we revealed that this fusion promotes lung SCC progression through regulating a SIRT6-ERK1/2-MMP1 signaling axis. Targeted therapy toward ERK1/2 activation using selumetinib provides an efficient means for treatment of lung SCC with the TRA2B-DNAH5 fusion. This study might hold great promise for therapeutic strategy development toward lung SCC patients harboring the TRA2B-DNAH5 fusion. Indeed, MMP1 seems to be a promising target for the treatment of lung SCC patients harboring TRA2B-DNAH5 fusion. Marimastat is a broad-spectrum inhibitor of MMPs which has high inhibitory effect on MMP1 (IC50 value is 5 nM for MMP1). We therefore tested the therapeutic effect of marimastat upon the growth of CRL-5889 xenograft tumors with TRA2B-DNAH5 fusion expression. To our disappointment, marimastat treatment failed to inhibit tumor growth (Supplementary information, Figure S16). This is somehow consistent with clinical trial results of marimastat46 and the lack of therapeutic efficacy might be due to non-specific targeting of multiple MMPs with counteracting function.
As we know, most oncogenic gene fusions are kinases, and only a few are transcriptional factors. Interestingly, the TRA2B-DNAH5 fusion is different from traditional oncogenic gene fusions, and the core component of TRA2B-DNAH5 fusion is DNAH5, which is an axonemal dynein. We find that TRA2B-DNAH5 fusion interacts with SIRT6 and inhibits its nuclear translocation, which leads to upregulation of the expression of multiple downstream target genes including TIAM1, SUV420H1, RIN1 and several IGF signaling-related genes, and then promotes ERK1/2 activation and MMP1 expression. Although it remains attractive to find a better way to target MMP1, our data show that selumetinib can efficiently inhibit the growth of lung SCC with TRA2B-DNAH5 fusion in xenograft assay. Selumetinib (AZD-6244; ARRY-142886), a MEK1/2 inhibitor designed by AstraZeneca, can inhibit ERK1/2 activity through targeting MEK1/247. Moreover, selumetinib was approved by the US Food and Drug Administration for the treatment of Uveal melanoma in 2015, and now this drug is under phase 3 clinic trial for the treatment of metastatic NSCLC. Our data indicate that selumetinib serves as a promising drug for those lung SCC patients harboring the TRA2B-DNAH5 fusion, which might help develop targeted therapy and precision medicine for lung cancer treatment.
Materials and Methods
Clinical specimen collection
This study was approved by the institutional review board of Fudan University Shanghai Cancer Hospital, Shanghai, China. All participants gave written informed consent. The 119 lung ADC and 163 SCC specimens with paired pathological normal lungs were collected from July 2007 to June 2011. Fresh surgical specimens were snap-frozen and stored in liquid nitrogen upon resection until use. The pathology of each tumor sample was determined by pathologists. All these specimens were with a minimum of 70% of tumor cellularity, and all patients did not receive neoadjuvant chemotherapy. Total RNA was extracted from tissues with Trizol Reagent (Invitrogen) according to the manufacturer's instruction.
Exon array-based fusion gene detection and gene expression profiling analyses
The method for fusion gene detection has been established in our previous study11. Fusion gene prediction was based on the Affymetrix Exon 1.0 dataset. For the microarray data analysis, we applied the Robust Multichip Average (RMA) method to perform background correction, normalization and exon-level probe set summarization. Exon-level expression values were then normalized across the samples, which allowed subsequent computational identification of breakpoints along the exons positioned in the chromosome order. To identify fusion genes characterized by divergent expression between the 5′ and 3′ sides partitioned by the breakpoint, a Student's t-test was used to evaluate each sample for such expression patterns (single-sample significance) and then an overall asymptotic significance was calculated by summarizing the t-statistics into a dataset-wise statistic Sg:

Here, k is the probe set number designed for gene g in the Exon 1.0 microarray, and n is the sample number. Note, the above computations require an exhaustive screening for all possible breakpoints in the gene under analysis, which would turn out to be a computationally expensive procedure when applied to whole-genome screen. To accelerate the screening process, we simplified the procedure by taking the middle point of probe sets for a gene as an initial breakpoint to identify candidate genes which were then sent for exact analysis aiming at predicting the optimal breakpoint.
Expression profiling was performed on Affymetrix Prime Preview Chips. After preprocessing, normalization and annotation, differential expressions were evaluated with fold change (fold > 1.5) and t-test (P < 0.05), and functional enrichment was analyzed with Fisher's exact test on KEGG pathways and Gene Ontology (GO) gene sets. With PCA, expression data composed of a set of differentially expressed genes was reduced in dimension to a few principal components (PCs), and contributions of those original genes to the three predominant PCs were assessed and visualized.
Molecular analysis for gene fusions
5′ RACE was performed using SMARTer™ RACE cDNA Amplification Kit from Clontech Laboratories (Mountain View, CA, USA) according to the manufacturer's instructions. Briefly, 1 μg RNA extracted from lung tumor specimens was reverse-transcribed using primers 5′-RACE CDS primer A and SMARTer II A Oligonucleotide supplied by SMARTer™ RACE cDNA Amplification Kit. PCR was performed with TIE1 gene-specific primer 5′-CACCACAGCCACCATCAGAACCAGAG-3′ or DNAH5 gene-specific primer 5′-TCCACGGCTTTGTTCAGGGTCTGCTGTA-3′ in conjunction with RACE universal primer A mix (UPM): 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′ and 5-CTAATACGACTCACTATAGGGC-3′. PCR products were purified for direct sequencing. Total RNA samples were also reverse-transcribed into single-stranded cDNA using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, EU) and used for detection of the potential TIE1 fusion. For detection of PTPRF-TIE1 fusion, the forward primer was 5′-CACTGGTGATGCTTGGTTTG-3′, and the reverse primer was 5′-GTGGTCTTCTCTGGCTCCAC-3′. For detection of TRA2B-DNAH5 fusion, the forward primer was 5′-AAGGAAGGTGCAAGAGGTTG-3′ and the reverse primer was 5′-AACTTCCACATCCAGCAACA-3′.
The PCR program for detection of PTPRF-TIE1 fusion and TRA2B-DNAH5 fusion is: 94 °C, 2 min; 94 °C, 15 s, 58 °C, 30 s, 72 °C, 30 s, 35 cycles; 72 °C, 1 min.
To identify the genomic breakpoint of TRA2B-DNAH5 fusion (Supplementary information, Data S1), genomic DNAs from lung cancer sample 207LC and paired normal lung tissue were used for long-range PCR using a series of primers (7 forward primers T1-T7 at TRA2B intron 1 with 1-2 kb intervals, and 3 reverse primers D1-D3 at DNAH5 intron 17 with 1-2 kb intervals).
T1: 5′-GCGGCGAGCAGAACTACGGCGAGCG-3′;
T2: 5′-ACTGCAAAGTAAGCGATTTTAAAGT-3′;
T3: 5′-GTTAATGGGATTTGCCTTACATAGC-3′;
T4: 5′-TGGTTGACTTGATTTTGTCTTTTTA-3′;
T5: 5′-ACACATATCAAAAATAATACCCAGT-3′;
T6: 5′-TGAAACACCCATATTCTGCTTTCAC-3′;
T7: 5′-TGAAACACCCATATTCTGCTTTCAC-3′;
D1: 5′-ACAGTTAGTGAATGTGAGGAGGAGG-3′;
D2: 5′-TTCAACCATTGTTTTGCTAATATCC-3′;
D3: 5′-ATTTGTGCACCATTTACACAAAGAT-3′.
The genomic breakpoint from lung cancer sample 207LC was amplified by the primer pairs T1 + D2, T1 + D3, T2 + D2 and T2 + D3. PCR products were visualized by 1.5% agarose gel electrophoresis, and the exact location of the breakpoint was obtained by regular Sanger sequencing.
Cell culture, plasmid construction and lentivirus infection
Ink4a−/− MEFs, HEK-293T and Beas2B cells were cultured in DMEM (Hyclone) with 10% fetal bovine serum (FBS). CRL-5889 and HTB-58 cells were cultured in RPMI-1640 (Hyclone) with 10% FBS.
PTPRF-TIE1 fusion with C-terminally fused Myc tag or TRA2B-DNAH5 fusion with C-terminally fused 6× Myc tag was cloned into pCDH-CMV-EF1-Puro (Systems Biosciences). SIRT6 with C-terminally fused 3× Flag tag was cloned into pCDH-CMV-EF1-CoGFP (Systems Biosciences). The shRNAs toward human MMP1 and SIRT6 were cloned into pLKO.1 vector (Addgene) with the AgeI/EcoRI sites. The target sequences of shRNAs were as follows:
shMMP1-1: 5′-CTTGAAGCTGCTTACGAATTT-3′
shMMP1-2: 5′-GCTAACCTTTGATGCTATAAC-3′
shMMP1-3: 5′-GTTTGTGGCTTATGGATTCAT-3′
shSIRT6-1: 5′-ACGGGAACATGTTTGTGGAAG-3′
shSIRT6-2: 5′-CAGTACGTCCGAGACACAGTC-3′
shSIRT6-3: 5′-CAAGTTCGACACCACCTTTGA-3′
Lentiviral package and infection was done as follows: HEK-293T cells were co-transfected with pCDH or pLKO.1 constructs and packaging plasmids. The progeny viruses released from HEK-293T cells were filtered, collected and used to infect cells. Stable cell lines were selected and maintained in 2 μg/ml puromycin.
Quantitative real-time PCR
Total RNA was extracted from cells and xenograft tumors with Trizol Reagent (Invitrogen) according to the manufacturer's instructions. Total RNAs were also reverse-transcribed into single-stranded cDNA using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, EU). cDNAs were subjected to quantitative real-time PCR with gene-specific primers on 7500 Fast Real-Time PCR System (Applied Biosystems) using SYBR Green Master PCR mix (Invitrogen). GAPDH (human), 16S RNA (human) or β-actin (mouse) served as internal control. Primers used for real-time PCR analyses were listed in Supplementary information, Table S5.
Western blot analysis
Stable cell lines or xenograft tumors were harvested for western blotting. Protein samples were probed with specific antibodies against MMP1 (Proteintech, 10371-2-AP, 1:1 000), phospho-ERK1/2 (T202/Y204, Cell Signaling, #9106S, 1:1 000), total ERK1/2 (Cell Signaling, #9102, 1:1 000), phospho-CaMKIIα (T286, Santa Cruz Biotechnology, #SC-12886-R, 1:1 000), phospho-PKAα/β (T197, Bioworld, BS4345, 1:1 000), phospho-PKCα/β (T648/641, Cell Signaling, #9375S, 1:1 000), phospho-PKCδ/θ (S643/676, Cell Signaling, #9376, 1:1 000), phospho-STAT1 (Y701, Cell Signaling, #9167S, 1:1 000), phospho-STAT2 (Y690, Cell Signaling, #4441S, 1:1 000), phospho-JNK (T183/Y185, Cell Signaling, #9255, 1:1 000), SIRT6 (Sangon, #D121220, 1:500), IGF2R (Sangon, #D161777, 1:500), IGF1R (Sangon, #D151673, 1:500), phospho-IGF1R (Absci, #AB11716, 1:500), MEK1/2 (Cell Signaling, #9154, 1:1 000), phospho-MEK1/2 (Cell Signaling, #8727, 1:1 000) or β-actin (Sigma, #A2228, 1:5 000). Protein expression was assessed by Pierce ECL Western Blotting Substrate (Thermo Scientific, IL, USA) and exposure to X-ray film (Eastman Kodak, NY, USA). The film was scanned, and the digitalized images were quantified by densitometry.
IP-MS analyses and Co-IP assay
CRL-5889 cells with or without 6× Myc-tagged TRA2B-DNAH5 fusion expression were lysed and the supernatants were incubated with anti-Myc antibody and protein G agarose (Invitrogen). The immune complexes were subjected to trypsin digestion and mass spectrometry analysis48. All mass spectrometric data were acquired using Xcalibur software and analyzed using MaxQuant 1.3.0.5 against the human International Protein Index (IPI) database (version 3.68).
HEK-293T cells were transfected with the indicated plasmids using calcium transfection. Cells were lysed and the supernatants were incubated with anti-Myc and anti-FLAG antibodies and protein G agarose (Invitrogen). The immune complexes were subjected to SDS-PAGE, and analyzed by western blot.
Immunostaining
Immunostaining was performed as previously described49. The antibodies used were p-ERK1/2 (T202/Y204, Santa Cruz Biotechnology, sc-16982-R, 1:100), MMP1 (Proteintech, 10371-2-AP, 1:500), Ki67 (Novocastra, NCL-KI67p, 1:500). The proliferation rate was evaluated by counting Ki67-positive nuclear staining at high-power field (HPF) for more than 30 fields for each group.
Cell proliferation assay and soft agar colony formation assay
For cell proliferation assay, cells were seeded in 96-well plates (Nest) and the viability of cells was measured at the indicated time points. For Beas2B and CRL-5889 cells, at different time points (Days 1-5), 20 μl MTT solution (5 mg/ml, Sigma) was added into each well. Cells were incubated at 37 °C for 4 h. Then the medium was removed and 100 μl DMSO was added to each well to solubilize the formazan crystals. Absorbance was measured at dual wave length mode (570 nm and 630 nm) using a Microplate Reader (Thermo Scientific, MULTISKAN MK3, USA).
For soft agar assay, 5 000 cells were plated in 0.4% top agarose on top of 1% agarose base supplemented with complete medium in 6-well plates. After 3 weeks, cells were stained with 0.005% crystal violet and colonies (>1 mm in diameter) were counted. All experiments were performed in triplicates.
Cell transformation assay
Ink4a−/− MEFs transfected with TRA2B-DNAH5 or CCDC6-RET fusion were cultured in DMEM with 10% FBS until 100% confluence. MEFs were changed with fresh medium every 3 days until the foci analyses using 0.005% crystal violet staining.
Xenograft assay and drug treatment
CRL-5889 or HTB-58 cells with or without TRA2B-DNAH5 fusion expression were subcutaneously transplanted into nude mice. Selumetinib were given through gavage at 50 mg/kg twice a day, and marimastat was given through intraperitoneal injection at 18 mg/kg once a day. Tumor volume was monitored twice a week, and mice were sacrificed for further molecular and pathological analysis.
Luciferase reporter assay
Six reporters including CRE, SRE, NF-κB, GRE, HSE, and AP1 are used in this study. HEK-293T cells were seeded in 24-well plates. Luciferase reporter and the indicated plasmids were co-transfected. Luciferase activities were measured 48 h after transfection using Dual-Luciferase Assay kit (Promega) on GloMax 20/20 luminometer (Promega) following the manufacturer's instructions. pRL-TK was co-transfected as internal control. Experiments were conducted in triplicates and repeated at least 3 times.
Statistical analysis
All statistical analyses were carried out using the SPSS 13.0 statistical software package. Data were analyzed by Student's t-test (two tailed). P < 0.05 was considered to be significant. Error bars represent SEM.
Data access
Expression profile data for CRL-5889 xenograft tumors and Affymetrix Exon 1.0 array data for human NSCLC and paired normal lungs from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE74046, GSE74095, GSE74115, GSE74116 and GSE74117.
Author Contributions
H Ji, F Li, Z Fang and J Zhang conceived the study. H Ji supervised the study. F Li and J Zhang performed the experiments. Z Fang contributed to the bioinformatics analysis. H Liu, J Xia, C Guo, Z Qin, FM Li, X Han, YT Wang, Y Feng, Y Wang, W Zhang, Z Wang and Y Jin provided technical help and helpful discussion. W Wei provided materials and helpful discussion. H Zhu, C Li and R Zeng helped with MS experiment and analysis. H Chen and Y Sun provided clinical samples. H Ji, F Li, J Zhang and Z Fang wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
Siegel R, Ma J, Zou Z, Jemal A . Cancer statistics. CA Cancer J Clin 2014; 64:9–29.
Herbst RS, Lippman SM . Molecular signatures of lung cancer — toward personalized therapy. N Engl J Med 2007; 356:76–78.
Riely GJ, Marks J, Pao W . KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 2009; 6:201–205.
Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 2004; 101:13306–13311.
Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304:1497–1500.
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350:2129–2139.
Ramos AH, Dutt A, Mermel C, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther 2009; 8:2042–2050.
Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov 2011; 1:78–89.
Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448:561–566.
Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007; 131:1190–1203.
Li F, Feng Y, Fang R, et al. Identification of RET gene fusion by exon array analyses in “pan-negative” lung cancer from never smokers. Cell Res 2012; 22:928–931.
Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012; 18:382–384.
Ju YS, Lee WC, Shin JY, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res 2012; 22:436–445.
Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012; 18:375–377.
Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012; 18:378–381.
Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013; 19:1469–1472.
Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013; 3:636–647.
Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014; 4:415–422.
Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013; 368:2385–2394.
Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014; 371:2167–2177.
Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014; 371:1963–1971.
Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013; 3:630–635.
Li F, Feng Y, Fang R, et al. Identification of RET gene fusion by exon array analyses in “pan-negative” lung cancer from never smokers. Cell Res 2012; 22:928–931.
Pao W, Girard N . New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011; 12:175–180.
Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010; 28:4616–4620.
Li C, Fang R, Sun Y, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One 2011; 6:e28204.
Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res 2009; 7:1466–1476.
Seegar TC, Eller B, Tzvetkova-Robev D, et al. Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol Cell 2010; 37:643–655.
Foley CJ, Kuliopulos A . Mouse matrix metalloprotease-1a (Mmp1a) gives new insight into MMP function. J Cell Physiol 2014; 229:1875–1880.
Kim CH, Park YG, Noh SH, Kim YK . PGE2 induces the gene expression of bone matrix metalloproteinase-1 in mouse osteoblasts by cAMP-PKA signaling pathway. Int J Biochem Cell Biol 2005; 37:375–385.
Sokolova O, Vieth M, Naumann M . Protein kinase C isozymes regulate matrix metalloproteinase-1 expression and cell invasion in Helicobacter pylori infection. Gut 2013; 62:358–367.
Zhang W, Chen DQ, Qi F, Wang J, Xiao WY, Zhu WZ . Inhibition of calcium-calmodulin-dependent kinase II suppresses cardiac fibroblast proliferation and extracellular matrix secretion. J Cardiovasc Pharmacol 2010; 55:96–105.
Kim HH, Shin CM, Park CH, et al. Eicosapentaenoic acid inhibits UV-induced MMP-1 expression in human dermal fibroblasts. J Lipid Res 2005; 46:1712–1720.
Reunanen N, Li SP, Ahonen M, Foschi M, Han J, Kahari VM . Activation of p38α MAPK enhances collagenase-1 (matrix metalloproteinase (MMP)-1) and stromelysin-1 (MMP-3) expression by mRNA stabilization. J Biol Chem 2002; 277:32360–32368.
Korzus E, Nagase H, Rydell R, Travis J . The mitogen-activated protein kinase and JAK-STAT signaling pathways are required for an oncostatin M-responsive element-mediated activation of matrix metalloproteinase 1 gene expression. J Biol Chem 1997; 272:1188–1196.
Sebastian C, Zwaans BM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012; 151:1185–1199.
Zhang ZG, Qin CY . Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signal-regulated kinase signaling pathway. Mol Med Rep 2014; 9:882–888.
Kugel S, Feldman JL, Klein MA, et al. Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer. Cell Rep 2015; 13:479–488.
Sundaresan NR, Vasudevan P, Zhong L, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med 2012; 18:1643–1650.
Mertens AE, Roovers RC, Collard JG . Regulation of Tiam1-Rac signalling. FEBS Lett 2003; 546:11–16.
Eblen ST, Slack JK, Weber MJ, Catling AD . Rac-PAK signaling stimulates extracellular signal-regulated kinase (ERK) activation by regulating formation of MEK1-ERK complexes. Mol Cell Biol 2002; 22:6023–6033.
Soto E, Yanagisawa M, Marlow LA, Copland JA, Perez EA, Anastasiadis PZ . p120 catenin induces opposing effects on tumor cell growth depending on E-cadherin expression. J Cell Biol 2008; 183:737–749.
Vougiouklakis T, Sone K, Saloura V, et al. SUV420H1 enhances the phosphorylation and transcription of ERK1 in cancer cells. Oncotarget 2015; 6:43162–43171.
Hunker CM, Giambini H, Galvis A, et al. Rin1 regulates insulin receptor signal transduction pathways. Exp Cell Res 2006; 312:1106–1118.
Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res 2014; 20:4107–4114.
Sparano JA, Bernardo P, Stephenson P, et al. Randomized phase III trial of marimastat versus placebo in patients with metastatic breast cancer who have responding or stable disease after first-line chemotherapy: Eastern Cooperative Oncology Group trial E2196. J Clin Oncol 2004; 22:4683–4690.
Yeh TC, Marsh V, Bernat BA, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res 2007; 13:1576–1583.
Wisniewski JR, Zougman A, Nagaraj N, Mann M . Universal sample preparation method for proteome analysis. Nat Methods 2009; 6:359–362.
Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007; 448:807–810.
Acknowledgements
We thank Drs Lei Zhang, Xiumin Yan, Zhuang Wei, Shuai Wu, Lin Pan and Liang Dong for technical supports. We thank Dangsheng Li for the critical reading of the manuscript and suggestions. We also thank all of our lab members for helpful discussion. This work was supported by the Ministry of Science and Technology of China (2012CB910800), the National Natural Science Foundation of China (81430066, 81402276, 81402371, 81401898, 81402498, 81101583, 81372509, 31370747 and 81325015), the Science and Technology Commission of Shanghai Municipality (12JC1409800, 15XD1504000), the “Cross and cooperation in science and technology innovation team” program, the Chinese Postdoctoral foundation (2014M561536, 2013T60476), the Shanghai Postdoctoral Foundation (14R21411400), and the Shanghai Institutes for Biological Sciences (2013KIP303, 2013KIP102, 2014KIP304).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
409 potential gene fusions or isoforms in ADCs. (XLS 53 kb)
Supplementary information, Table S2
284 potential gene fusions or isoforms in SCCs. (XLS 43 kb)
Supplementary information, Table S3
Clinical features of 5 lung SCC patients harboring TRA2B-DNAH5 fusion (PDF 8 kb)
Supplementary information, Table S4
Identification of TRA2B-DNAH5 fusion interacting proteins IP-MS analyses. (XLS 24 kb)
Supplementary information, Table S5
The list of primers used for real-time PCR quantification in this study. (PDF 19 kb)
Supplementary information, Figure S1
Identification of TIE1 fusion in human lung ADC. (PDF 172 kb)
Supplementary information, Figure S2
Identification of genomic breakpoint of TRA2B-DNAH5 fusion. (PDF 153 kb)
Supplementary information, Figure S3
Sanger sequencing confirms the TRA2B-DNAH5 fusion in another 4 human lung SCC specimensat RNA level. (PDF 122 kb)
Supplementary information, Figure S4
The TRA2B-DNAH5 fusion promotes cell transformation. (PDF 51 kb)
Supplementary information, Figure S5
Immunohistochemical studies of CRL-5889 xenograft tumors. (PDF 154 kb)
Supplementary information, Figure S6
Confirmation of MMP1 up-regulation and ERK1/2 activation in cells and xenograft tumors. (PDF 166 kb)
Supplementary information, Figure S7
Up-regulated MMP1 expression mediates the promotive function of TRA2B-DNAH5 fusion in cell invasion and migration. (PDF 142 kb)
Supplementary information, Figure S8
Knockdown of MMP1 significantly inhibits the tumor-promotivefunction of the TRA2B-DNAH5 fusion in CRL-5889 xenograft assay. (PDF 114 kb)
Supplementary information, Figure S9
Human lung SCC specimens harboring the TRA2B-DNAH5 fusion are stained positive for p-ERK1/2 and MMP1. (PDF 172 kb)
Supplementary information, Figure S10
TRA2B-DNAH5 fusion is mainlylocated at cytoplasm. (PDF 52 kb)
Supplementary information, Figure S11
CHIP-Seq data reveals the binding of SIRT6 on TIAM1, SUV620H1 and RIN1 genes. (PDF 71 kb)
Supplementary information, Figure S12
IGF1R may be involved in the activation of MEK1/2 and up-regulation of MMP1 by TRA2B-DNAH5 fusion. (PDF 75 kb)
Supplementary information, Figure S13
Human lung SCC cells expressing the TRA2B-DNAH5 fusion are more sensitive to selumetinib treatment. (PDF 118 kb)
Supplementary information, Figure S14
Selumetinib treatment inhibits ERK1/2 activity and MMP1 levels in CRL-5889 xenograft tumors with TRA2B-DNAH5 fusion expression. (PDF 164 kb)
Supplementary information, Figure S15
Selumetinib treatment doesn't cause a significant weight loss in mice. (PDF 112 kb)
Supplementary information, Figure S16
Marimastat treatment has no inhibitory effect on tumor growth of CRL-5889 xenograft tumors. (PDF 61 kb)
Supplementary information, Data S1
CDS sequence and amino acids sequence of TRA2B-DNAH5 fusion. (PDF 44 kb)
Rights and permissions
About this article
Cite this article
Li, F., Fang, Z., Zhang, J. et al. Identification of TRA2B-DNAH5 fusion as a novel oncogenic driver in human lung squamous cell carcinoma. Cell Res 26, 1149–1164 (2016). https://doi.org/10.1038/cr.2016.111
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2016.111
Keywords
This article is cited by
-
Switched alternative splicing events as attractive features in lung squamous cell carcinoma
Cancer Cell International (2022)
-
T cell differentiation protein 2 facilitates cell proliferation by enhancing mTOR-mediated ribosome biogenesis in non-small cell lung cancer
Discover Oncology (2022)
-
Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression
Scientific Reports (2021)
-
Identification of a novel gene signature for the prediction of recurrence in HCC patients by machine learning of genome-wide databases
Scientific Reports (2020)