Abstract
Telomere maintenance is critical for genome stability. The newly-identified Ctc1/Stn1/Ten1 complex is important for telomere maintenance, though its precise role is unclear. We report here that depletion of hStn1 induces catastrophic telomere shortening, DNA damage response, and early senescence in human somatic cells. These phenotypes are likely due to the essential role of hStn1 in promoting efficient replication of lagging-strand telomeric DNA. Downregulation of hStn1 accumulates single-stranded G-rich DNA specifically at lagging-strand telomeres, increases telomere fragility, hinders telomere DNA synthesis, as well as delays and compromises telomeric C-strand synthesis. We further show that hStn1 deficiency leads to persistent and elevated association of DNA polymerase α (polα) to telomeres, suggesting that hStn1 may modulate the DNA synthesis activity of polα rather than controlling the loading of polα to telomeres. Additionally, our data suggest that hStn1 is unlikely to be part of the telomere capping complex. We propose that the hStn1 assists DNA polymerases to efficiently duplicate lagging-strand telomeres in order to achieve complete synthesis of telomeric DNA, therefore preventing rapid telomere loss.
Similar content being viewed by others
Introduction
Human telomere sequences are composed of several kilobases of (TTAGGG)n repeats followed by long single-stranded (ss) overhangs at the 3′ end of the G-rich sequences, called G-overhangs. Maintaining the integrity of telomeres is crucial for safeguarding genome stability. Telomere stability is maintained by a number of proteins binding to telomeres, including the six-member shelterin complex (TRF1, TRF2, POT1, TPP1, TIN2, and RAP1)1. Deficiencies in these proteins “uncap” chromosome ends and activate DNA damage response (DDR) pathways, leading to inappropriate chromosome end-to-end fusions or recombinations that drive genome instability.
Recently, a trimeric protein complex comprising Ctc1, Stn1, and Ten1 (the CST complex), has been identified as a new regulator of telomere integrity2,3,4,5, adding more complexity of telomere end protection. Ctc1 and Stn1 were initially identified as co-factors of DNA polymerase α (polα)-primase complex and regulate DNA replication in mammalian cells6. CST binds to ss telomeric DNA, although the binding lacks sequence specificity in vitro2. Deficiency of Ctc1 or Stn1 lengthens G-overhangs2,3,4. Genetic variants of Stn1 are associated with increased telomere length in leukocytes7 and expressing a deletion mutant of Stn1 in cancer cells causes telomere lengthening5, suggesting that Stn1 may be involved in telomere length regulation. Recently, mutations in Ctc1 have been isolated from patients with telomere disorder dyskeratosis congenita (DC) and patients with Coats Plus (CP) syndrome, a complex genetic disease clinically related to DC8,9,10. Shortened telomeres were observed in both CP and DC patients carrying Ctc1 mutations8,10, suggesting that the underlying cause for CP may be related to defective telomere maintenance induced by Ctc1 mutations. Though it has been shown that transient depletion of Ctc1 or Stn1 in HeLa leads to telomere instability3,4, the precise role of human CST complex is largely unknown. How CST coordinates with shelterins to protect telomeres and whether the three components of the CST complex function equally in protecting chromosome ends are central questions in the telomere biology field.
In yeasts and plants, homologs of CST are critical for ensuring telomere stability3,11,12,13,14,15,16,17,18,19,20,21,22. In budding yeast, the Cdc13/Stn1/Ten1 complex associates exclusively with the G-overhang and plays important roles in restricting excessive C-strand resection from telomere ends, mediating C-strand fill-in, and regulating telomerase activity14,15,16,17,18,19,20,21,22. In fission yeast, orthologs of Stn1 and Ten1 have been identified and they are essential for cell growth. Deletion of Stn1 or Ten1 causes complete telomere loss and intrachromosomal end fusions11. In plants, Ctc1/Stn1/Ten1 plays a major role in chromosome end protection. Ctc1- and Stn1-null mutants in Arabidopsis suffer dramatic telomere shortening, end-to-end chromosomal fusions, increased G-overhangs, and elevated extrachromosomal telomeric circles3,12.
High fidelity and complete replication of telomeric DNA is critical for maintaining telomere stability, in particular for preventing rapid telomere loss. Telomeres possess special features, such as the repetitive DNA sequence, the four-stranded G-quadruplex structure, the unique telomere capping structure, and abundant telomere-binding proteins. These features pose challenges to replication machinery, and replication forks are vulnerable to stalling at telomeric tracts23,24. This is validated by recent reports demonstrating that telomeres resemble common fragile sites and efficient replication of telomeric DNA requires assistance from telomere-binding proteins as well as associated factors25,26,27,28,29,30.
In this study, we characterized the effects of Stn1 deficiency on telomere maintenance in several aspects including telomeric DNA replication, C-strand synthesis, and telomere end resection in human cells. We report that loss of Stn1 delays BrdU incorporation into daughter telomeres during replication, accumulates excessive G-rich ssDNA predominantly at telomeres replicated by lagging-strand synthesis, and increases telomere fragility. These phenotypes suggest an essential role of hStn1 in promoting efficient replication of telomeres. Examination of the cell cycle-regulated G-overhang dynamics shows that depletion of Stn1 delays and compromises C-strand synthesis that occurs in the late S/G2 phase, leading to persistent lengthening of G-overhangs. We have also found that downregulation of Stn1 leads to rapid telomere shortening and early cellular senescence in telomerase-negative human fibroblasts. Interestingly, Stn1 deficiency elevates and prolongs polα association with telomeres, suggesting that Stn1 is dispensable for recruiting polα to chromosome ends and may modulate the synthesis activity of polα at telomeres. Collectively, our results suggest that the primary function of Stn1 is to promote efficient replication of lagging-strand telomeres. These findings provide mechanistic insights into the cellular roles of human Stn1 in telomere maintenance.
Results
Deficiency in Stn1 produces ss DNA predominantly at G-rich strand in both telomerase-positive and -negative cells
We previously reported that acute knockdown (KD) of Stn1 induced DDR, G-overhang elongation, and cell death in HeLa cells 72 h after transient siRNA transfection4. We now used shRNA to stably deplete Stn1. Cells were infected with retrovirus containing Stn1 shRNA, selected, and puromycin-resistant cells were pooled immediately after selection. Western blot showed ∼90% Stn1 protein was removed by shRNA (Figure 1A and Supplementary information, Figure S1B). To ensure that the anti-Stn1 antibody was able to detect Stn1, we purified recombinant His6-Stn1 from E. coli (Supplementary information, Figure S1A) and tested whether the antibody recognized purified His6-Stn1 protein. Anti-Stn1 antibody specifically recognized Stn1 protein (Supplementary information, Figure S1B), suggesting that the detected band was indeed Stn1.
Elongated G-overhang and persistent ExoI-resistant ss DNA at G-rich strand caused by Stn1 depletion in both telomerase-positive and -negative cells. (A) Western blot of Stn1 KD in HeLa and BJ cells, as well as expression of FLAG-tagged RNA-resistant Stn1 (FLAG-MM-Stn1). Cells were infected with retroviral shRNA and selected for puromycin resistance. One day after selection completed, cells were pooled and used for DNA isolation and western blot. Luc: shRNA targeting luciferase. (B) In-gel hybridization of HeLa and BJ cells expressing shStn1. MM-Stn1: RNAi-resistant Stn1 containing mismatch in RNAi targeting region. (C) Quantitation of relative abundance of ExoI-resistant ss G-rich DNA and ExoI-sensitive G-overhangs in HeLa and BJ cells. Signals were normalized to Alu signals. Two-tailed t-test was used to calculate statistical significance. Results were from 5 independent experiments. Error bars: S.E.M. (D) In-gel hybridization with C-rich or G-rich telomeric probe in HeLa cells expressing shRNA. RecJf is a 5′→3′ exonuclease that specifically removes 5′ overhangs.
In contrast to acute KD-induced cell death, stable depletion of Stn1 in HeLa had undetectable effect on cell growth. We repeated acute Stn1 KD using the same siRNA sequences as previously described and observed similar cell death in HeLa cells (data not shown), suggesting that acute Stn1 KD had a more severe impact on cell growth. Although the underlying reason for cell death induced by acute siRNA KD remains to be investigated, it is possible that during retroviral infection and selection, stable shRNA-expressing clones that had lower level of KD were preferably selected for survival.
To determine the functions of Stn1 at telomeres, we first applied non-denaturing in-gel hybridization assay to analyze the integrity of telomeric DNA in Stn1-KD cells (Figure 1). Genomic DNA isolated from cells was treated with or without Exonuclease I (ExoI) prior to hybridization, which specifically removes 3′ ss G-overhangs. Both telomerase-positive HeLa cells and telomerase-negative BJ fibroblasts showed similar increase in ExoI-resistant ss DNA as well as G-overhangs (ExoI-sensitive) (Figure 1B and 1C), suggesting that Stn1 has a general role in preserving telomeric DNA integrity regardless of telomerase activity. Accumulation of ss DNA was rescued when RNAi-resistant Stn1 (MM-Stn1) was expressed (Figure 1B and 1C), suggesting that the KD was specific.
To determine whether ss DNA was equally accumulated at both G- and C-rich strands, we then hybridized DNA to G-rich or C-rich probe under non-denaturing condition. We observed that hybridizing to C-rich probe gave rise to strong ExoI-resistant ss DNA signal, whereas G-rich probe produced minimal hybridization signal (Figure 1D). Our results suggest that ss DNA in Stn1-depleted cells predominantly locates at G-rich strand and gaps are present at C-rich strand.
Depletion of Stn1 increases fragile telomeres
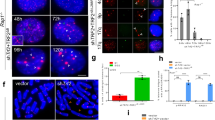
We suspected that the excessive ss DNA resulting from Stn1 deficiency might have stemmed from replication stress at telomeres. Under replication stress, mammalian telomeres resemble common fragile sites, represented as “broken” telomeres containing multiple-telomere signals at metaphase chromosomes27. We then expressed Stn1 shRNAs in HeLa 1.2.11, a cell line that contains 10-20 kb telomeres and is widely used in telomere FISH analysis. We found that the frequency of telomere breakage in Stn1-KD cells increased when metaphase chromosomes were hybridized to telomere probe (Figure 2A). To address whether the observed telomere breakage in Stn1-deficient cells resulted from fragile telomeres (FT), we treated HeLa 1.2.11 control and Stn1-KD cells with low concentration of aphidicolin, a drug that inhibits DNA polymerase α/δ and induces fragile site instability and breakage including FT. Aphidicolin treatment in control cells increased FT (Figure 2A), similar to what was reported27,29. Interestingly, aphidicolin treatment did not increase FTs in cells lacking Stn1 (Figure 2A). This suggests that Stn1 is likely in the same pathway as DNA polymerase α/δ in preventing FT induced by replication stress.
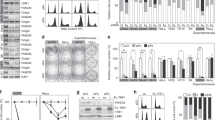
Stn1 deficiency impairs efficient replication of telomeres. (A) FTs induced by Stn1 KD. Metaphase chromosomes from HeLa 1.2.11 cells depleted of Stn1 were hybridized to PNA TTAGGG telomere probe. Representative telomere FISH images of shStn1 and control cells (shLuc) are shown. Three individual chromosomes are shown in amplified images to illustrate FTs (white arrows). Quantitation of FTs was performed by analyzing a total of > 75 metaphase spreads from each sample. At least 3 000 chromosomes were scored in each sample. Two-tailed t-test was used to calculate statistical significance. Results were from three independent experiments. (B) In-gel hybridization of separated leading and lagging telomeres from asynchronous HeLa cells with C-rich probe. Cells were labeled with BrdU continuously for 19 h and leading and lagging daughter telomeres were separated by CsCl gradient ultracentrifugation. (C) Quantitation of the abundance of ss G-rich DNA. Signals were normalized to the total telomere signals. Results were from three independent experiments. Error bars: S.E.M. (D) FACS analysis of DNA content in synchronized HeLa cells expressing shLuc and shStn1. Note that ∼10% cells failed to enter the cell cycle (Supplementary information, Figure S3B). (E) Separation of leading/lagging/unreplicated telomeres from cells collected at 6, 8, and 10 h after releasing from double thymidine block. Note the density shift of lagging telomere at 6 h (dashed lines) but not leading telomere in shStn1 cells. Results were from 6 independent experiments. P = 0.005. Two-tailed t-test was used to calculate statistical significance. (F) Percentage of unreplicated telomeres among the total telomeres at 6, 8, and 10 h after release. Two-tailed t-test was used to calculate statistical significance. P = 0.015 at 6 h, P = 0.019 at 8 h. Results were from at least 4 independent experiments. Error bars: S.E.M.
Since excessive ss DNA may potentially initiate recombination between telomeric DNA from sister chromatids, we then analyzed whether there was an increase in telomere recombination by measuring telomere sister chromatid exchange (T-SCE). No increase in T-SCE was found in Stn1-KD HeLa 1.2.11 cells (Supplementary information, Figure S2). Therefore, depletion of Stn1 alone did not increase recombination at telomeres. In addition, no end-to-end fusions were observed in Stn1-KD HeLa 1.2.11 (data not shown).
Stn1 promotes efficient replication of lagging-strand telomeres
Given that Stn1 deficiency increased FTs, we reasoned that Stn1 deficiency caused telomere replication defects. The two strands of telomeric DNA are duplicated by leading- and lagging-strand synthesis, respectively. Since G-strand is the parental strand for lagging-strand synthesis, increased G-rich ss DNA (Figure 1) indicates that lagging telomere replication may be disturbed. To firmly determine this, we separated leading and lagging telomeres as described previously4,31. By hybridizing separated daughter telomeres to C-rich probe, we found that Stn1 KD significantly increased ExoI-resistant ss DNA at lagging daughter telomeres but not at leading daughter telomeres (Figure 2B and 2C, ExoI-resistant signal). Thus, Stn1 deficiency primarily disrupted the integrity of lagging telomeres.
We hypothesized that Stn1 KD might impair efficient replication of lagging-strand synthesis. To test this, we then measured the efficiency of BrdU incorporation into daughter telomeres during replication. Cell cycle progression was largely unaltered in Stn1-depleted cells (Figure 2D), allowing for direct comparison of DNA synthesis in Stn1-KD and control cells. Cells were synchronized at G1/S boundary, released into S phase in the presence of BrdU, and collected at the mid to late S (6 h), the late S/G2 phase (8 h), and G2 phase (10 h). Replicated daughter telomeres and unreplicated telomeres were then separated on CsCl density gradient (Figure 2E and Supplementary information, Figure S3A). We noticed that telomere replication was delayed in Stn1-depleted cells, as indicated by slower disappearance of unreplicated telomeres from mid S to G2 phase (6 to 10 h) (Figure 2F), suggesting that telomere replication was less efficient or delayed. In addition, the density of lagging daughter telomeres in Stn1-KD cells was slightly lower than that in control, whereas such density shift was undetectable for leading telomeres (Figure 2E). Since the efficiency of BrdU incorporation determines the density of daughter telomeres, these results indicate that Stn1 KD led to inefficient replication of lagging daughter strands. The undisturbed leading-strand telomere density also argues against the possibility that the decreased density in lagging telomeres might be caused by any undetectable delays in cell cycle progression in Stn1-KD cells. Taken together, our data suggest that Stn1 likely promotes efficient replication of lagging-strand telomeres.
Deficiency of Stn1 induces rapid telomere shortening, early senescence and DDR in telomerase-negative fibroblasts
We noticed that Stn1 deficiency did not induce telomere loss in HeLa 1.2.11 (Supplementary information, Figure S2B), and FT appeared to be the only chromosome defect in HeLa 1.2.11 expressing shStn1. It has been reported that replication stress induced by aphidicolin treatment does not shorten telomeres in telomerase-positive cancer cells, but instead leads to moderate telomere lengthening27. Similarly, stable KD of FEN1, a nuclease that is important for replicating lagging-strand telomeres, in telomerase-positive cancer cells induces FT but has no effect on cell growth32. Hence, it is possible that telomerase in cancer cells may prevent FT from being shortened. To determine the effects of Stn1 deficiency in telomerase-negative cells, we then depleted Stn1 in BJ fibroblasts. Stn1 KD in BJ induced early senescence and growth arrest (Figure 3A and Supplementary information, Figure S4A). We suspected that the growth arrest might be caused by p53- or Rb-mediated checkpoint signaling in response to DNA damage. We then used BJ cells expressing E6 and E7 human papillomavirus oncoproteins, expression of which abolishes the functions of p53 and Rb. Stable depletion of Stn1 in BJ/E6/E7 showed normal cell growth (Supplementary information, Figure S4B), allowing us to perform further analysis on telomeric DNA. DNA was isolated for STELA to detect critically short telomeres. About 3 PD after Stn1 KD, short STELA products (< 0.9 kb) accumulated (Figure 3B and 3C, PD42). Because STELA products include the ∼410 bp subtelomeric region, STELA products with < 0.9 kb suggested telomere repeat length of < 0.5 kb. Additionally, extremely short PCR products with the size of ∼500 bp appeared upon Stn1 depletion (Supplementary information, Figure S5A). This size suggests telomeric tract lengths of ∼100 bp.
Rapid telomere shortening, early senescence, and DDR induced by Stn1 depletion in telomerase-negative BJ fibroblasts. (A) Early senescence induced by Stn1 KD in BJ cells. BJ cells were infected with retrovirus containing shLuc or shStn1 at PD47. Senescence-associated β-galactosidase staining was performed 4 days after selection for puromycin resistance. (B) STELA for measuring short telomeres at XpYp chromosome after Stn1 KD. BJ/E6/E7 cells were infected with retrovirus containing shLuc or shStn1 at PD35 and then selected in puromycin for 5 days. Cells were at PD39 after selection was completed. Tel3 is the telorette ending with the sequence CTAACC-3′ and TelN represents STELA using the combination of the other five telorettes ending with the sequences CCCTAA-3′, CCTAAC-3′, TAACCC-3′ AACCCT-3′, and ACCCTA-3′. (C) Percentage of short XpYp telomeres with the length of < 0.5 kb. Two-tailed t-test was used to calculate statistical significance. Results were from 6 independent experiments. Error bars: S.E.M. (D) Signal free ends detected in BJ/E6/E7 (PD43) cells expressing shStn1. Representative telomere CO-FISH images of shStn1 and control cells (shLuc) are shown. Quantitation was performed by analyzing a total of > 50 metaphase spreads for each sample. At least 2 000 chromosomes were scored in each sample. Results were from two experiments. Error bars: S.D. (E) Chk2 and Chk1 phosphorylation was induced by Stn1 KD. L: Luc shRNA. S: Stn1 shRNA. UV: cells treated with UV irradiation. (F) γ-H2AX foci (red) in BJ/E6/E7 cells expressing shStn1 at PD50. Green: telomere signal from hybridization with telomere probe. A total of over 150 nuclei were analyzed for each sample. Cells containing ≥ 5 foci were counted as positive γ-H2AX staining. Error bars: S.D. (G) ss DNA was accumulated shortly after Stn1 KD. Genomic DNA from BJ/E6/E7 shortly after Stn1 shRNA expression (PD42) was hybridized in gel under native condition (native) to C-probe. The same gel was then denatured and hybridized to Alu probe.
Since Stn1 KD accumulates ExoI-resistant internal ss G-rich telomeric DNA, it is possible that telorettes may anneal to these ss regions to produce short STELA products. To distinguish whether the observed short telomeres were due to telomere shortening or resulted from telorettes ligating at internal ss telomeric sites, we then treated DNA with ExoI prior to ligation to remove G-overhangs. ExoI treatment completely removed STELA products in control, while some ExoI-resistant products remained in the Stn1-KD sample (Supplementary information, Figure S5A), indicating that a small portion of STELA products from Stn1-KD DNA were indeed from telorettes ligating at internal ss sites. However, their contribution to short telomeres was minimal (Supplementary information, Figure S5B). Therefore, the accumulated short STELA products in Stn1-KD cells resulted mainly from telorettes ligating at the ends of shortened telomeres.
We then used FISH to confirm the existence of short telomeres observed in STELA. Our results showed that Stn1 depletion led to an increase in signal free ends at both leading and lagging daughter telomeres (Figure 3D). Together with STELA, our results strongly indicate that Stn1 deficiency in telomerase-negative cells induces rapid telomere loss.
Following Stn1 KD, Chk2 phosphorylation and γ-H2AX foci were detected (Figure 3E and 3F), indicating the activation of ATM and DDR. Interestingly, the majority of γ-H2AX foci did not colocalize with telomere signal (Figure 3F). This could be because γ-H2AX was recruited to extremely short telomeres that were undetectable by FISH. However, given that CST binds to ss DNA in a sequence-independent manner2, it is likely that CST has a role in preserving the integrity of general DNA sequences. Stn1 deficiency may trigger damage at non-telomeric regions, which also contributed to the observed DDR. The role of CST in maintaining the integrity of non-telomeric DNA sequences remains to be elucidated.
Since accumulation of ss DNA can trigger DDR by ATR activation33,34, we next examined whether the excessive ss telomeric DNA in Stn1-KD cells triggered ATR activation by analyzing phosphorylation of its downstream target Chk1. Chk1 phosphorylation was nearly undetectable immediately following Stn1 KD (Figure 3E, PD39). In addition, no RPA foci or Rad51 binding to telomeres was observed by immunofluorescence (data not shown). Elevated Chk1 phosphorylation appeared at later PDs, concurrent with the appearance of Chk2 phosphorylation (Figure 3E, PD47). Non-denaturing in-gel hybridization analysis revealed that ss G-rich DNA already accumulated in early PDs shortly after shRNA expression (Figure 3G). The failure to observe immediate ATR activation and RPA binding to telomeres suggests that either the amount of ss DNA was insufficient to activate ATR, or RPA was precluded from binding to the accumulated ss DNA in Stn1-deficient cells. This is perhaps due to the presence of Pot1 (see below, Figure 6), which suppresses ATR activation at telomeres by preventing RPA binding to telomeres35,36,37,38,39.
Stn1 depletion resulted in increased and prolonged polα association with telomeres, but did not affect the association of Pot1 with telomeres. (A) FACS analysis of DNA content in synchronized HeLa cells expressing shLuc and shStn1. (B) Representative slotblot results from telomere ChIP of polα and Pot1. A: asynchronous cells. (C, D) Quantitation of ChIP of polα and Pot1. Results were from at least 3 independent experiments. Two-tailed t-test was used to calculate statistical significance. Error bars: S.E.M.
No significant randomization of the final nucleotide at C-strand was found (Figure 3B), consistent with our previous report4 and suggesting that Stn1 unlikely regulates end resection at C-strand.
Deficiency in Stn1 delays G-overhang lengthening in S phase and compromises C-strand fill-in in G2 phase
In the late S/G2 phase, polα mediates an extra DNA synthesis step at the end of telomeric C-strand, termed C-strand fill-in or C-strand synthesis, leading to G-overhang shortening4,40. To examine whether CST is important for C-strand fill-in, we monitored G-overhang shortening during late S/G2 in HeLa cells stably depleted of Stn1. HeLa cells were synchronized at G1/S boundary and released into S phase (Figure 4A). In control cells, G-overhangs shortened by ∼30 nt from late S/G2 (7 h) to the next G1 (12 h). In Stn1-KD cells, G-overhangs remained lengthened after G2 phase albeit there was a moderate shortening (Figure 4B and 4C), suggesting that the shortening of G-overhangs was compromised by Stn1 KD.
Stn1 deficiency delayed G-overhang lengthening in S phase and compromised G-overhang shortening in G2 phase. (A) FACS analysis of DNA content in synchronized HeLa cells expressing shLuc and shStn1. (B) G-overhang measurement of shLuc and shStn1 at 7 h (late S/G2) and 12 h (next G1) after releasing from doublethymidine block using the overhang protection assay. (C) Quantitation of mean overhang lengths. Two-tailed t-test was used to calculate statistical significance. Results were from at least 4 independent experiments. Error bars: S.E.M. (D) Non-denaturing in-gel hybridization of ss G-rich DNA with C-probe during the cell cycle. (E) Quantitation of (D). Results were from 3 independent experiments. Error bars: S.D.
We then determined whether CST controlled the cell cycle-regulated G-overhang dynamics (Figure 4D and 4E). In control cells, G-overhangs were lengthened during S phase, reaching maximum length at ∼8 h (late S/G2) and then shortened by ∼40% at the next G1 (14 h). In Stn1-KD cells, there was a delay in reaching the maximal G-overhang length (at 10 h instead of 8 h, Figure 4E), as well as a reduced G-overhang shortening in G2 (by ∼10%-15% from 10 h to 14 h). Taken together, our results suggest that Stn1 deficiency delayed G-overhang lengthening in S phase as well as compromised C-strand fill-in in G2 phase, both of which support our conclusion that Stn1 promotes efficient replication of C-strands.
Stn1 deficiency elongates G-overhangs at leading-strand telomeres in telomerase-positive cells but not in telomerase-negative fibroblasts
We noticed that Stn1 KD in HeLa cells increased the abundance of G-overhangs at both leading and lagging telomeres (ExoI-sensitive signal in Figure 2B and 2C). Direct measurement of G-overhang length using overhang protection assay41 confirmed this result (Figure 5A and 5B). The sizes of G-overhangs are known to be controlled by three mechanisms: 5′→3′ resection of C-strand, telomerase extension of G-strand, and C-strand fill-in. To determine whether Stn1 regulates G-overhang generation at leading and lagging telomeres in the same way in telomerase-negative cells, we performed the assays using telomerase-negative BJ/E6/E7 cells. Stn1 KD elongated lagging overhangs in BJ/E6/E7 cells, similar to that in HeLa cells (Figure 5C and 5D). In contrast, leading G-overhangs remained unchanged upon Stn1 depletion in BJ/E6/E7 (Figure 5C and 5D). Previously we have shown that C-strand fill-in occurs at lagging daughter telomeres but is absent at leading telomeres, and therefore G-overhangs at leading telomeres in telomerase-positive cells are generated by C-strand end resection and telomere extension4. Our results that Stn1 deficiency elongates leading overhangs in telomerase-positive cells but not in telomerase-negative cells imply that Stn1 may regulate telomerase activity. Moreover, the unchanged G-overhang sizes at leading daughter telomeres in BJ/E6/E7 suggested that Stn1 unlikely played a role in controlling end resection at C-strand. The lengthened overhangs at lagging telomeres in both HeLa and BJ/E6/E7 (Figures 2C and 5C, 5D, ExoI-sensitive signals) may be due to defective C-strand fill-in induced by Stn1 KD (see above).
Stn1 unlikely plays a role in restricting end resection at C-strand. (A) G-overhang measurement of leading and lagging telomeres from HeLa cells using the overhang protection assay. (B) Quantitation of mean overhang lengths from A. Results were from 3 independent experiments. Error bars: S.E.M. (C) In-gel hybridization of separated leading and lagging telomeres from BJ/E6/E7 cells with C-rich probe. (D) Quantitation of the abundance of ss G-rich DNA from C. Signals were normalized to the total telomere signals. Results were from 4 independent experiments. Error bars: S.E.M. Two-tailed t-test was used in all the above experiments to calculate statistical significance.
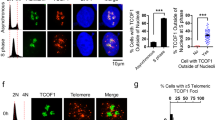
Stn1 deficiency results in persistent and increased association of polα with telomeres but does not alter Pot1 association with telomeres
Defects in replication could be due to defective recruitment of replication proteins to telomeres and/or compromised synthesis activities of these proteins upon encountering telomeric DNA. Based on the findings that Stn1 associates with both polα and shelterin component TPP15,42, it has been proposed that CST might recruit polα to telomeres. We found that Stn1 associated with polα throughout the cell cycle (Supplementary information, Figure S6), suggesting that the Stn1/polα interaction unlikely regulates polα recruitment to telomeres. We then used quantitative ChIP to determine the effect of Stn1 deficiency on polα association with telomeres. Surprisingly, upon depletion of Stn1, the telomeric association of polα was slightly enhanced in S phase and increased at late G2 phase (Figure 6B and 6C, 10 h). Such increase persisted throughout G2/M to the next G1 phase (12 h to 14 h) (Figure 6B and 6C). Although at present the underlying reason for this increased polα-telomere association is unclear, our data suggest that Stn1 is dispensable for polα association to telomeres. It is possible that Stn1 may affect polα's enzymatic activity at the telomeric region, as suggested by reports that Stn1 stimulates polα's DNA synthesis activity in vitro6,43.
We next determined whether the accumulated ss DNA and elongated G-overhangs in Stn1-deficient cells altered the association of another ss telomere-binding protein Pot1 to telomeres. ChIP analysis showed that Pot1 association with telomeres during the cell cycle remained largely unaltered upon Stn1 depletion (Figure 6B and 6D).
Discussion
We have found that deficiency of hStn1 impairs efficient replication of lagging-strand telomere DNA. As a result, downregulation of hStn1 accumulates ss G-rich DNA specifically at lagging-strand telomeres, increases telomere fragility, and delays synthesis of lagging-strand telomeres. Our findings establish that hStn1 protects the integrity of telomeres and prevents rapid loss of telomere repeats in the absence of telomerase activation. Given that Ctc1 and Stn1 stimulate polα's enzymatic activity6,43, we propose that the primary role of human CST in telomere protection is to facilitate the synthesis of telomere DNA chains on the lagging-strand templates, which possess natural hurdles such as G-quadruplexes that prevent efficient DNA replication. Because CST binding to DNA is sequence independent, it is likely that CST may have a role in general DNA synthesis, in particular at difficult-to-replicate regions.
While this manuscript was being revised, Gu et al.44 published a study demonstrating that mammalian Ctc1 promotes efficient replication of telomeric DNA and deletion of mammalian Ctc1 leads to telomere loss and stem cell failure. Absence of CTC1 induces progressive telomere loss and end-to-end fusions in late-passage MEFs. Acute deletion of CTC1 does not affect the localization of shelterin to telomeres, and DDR occurs only at chromosome ends devoid of telomeres44, suggesting that CTC1 deletion results in rapid telomere shortening. Our results of human Stn1 are largely consistent with mammalian CTC1 study. In addition, we provide direct evidence that Stn1 deficiency mainly affects lagging-strand telomere synthesis.
During replication, polα is brought to replication origins via interaction with Mcm10 and And-1/Ctf4 to initiate lagging-strand synthesis45,46,47. In the late S/G2 phase, polα is recruited back to telomeres48 (Figure 6), presumably for filling in C-strands. It is currently unknown how polα is recruited to telomere ends in late S/G2 phase. Previous findings that CST interacts with polα42 and TPP15 have led to the postulation that CST may recruit polα to telomeres. Surprisingly, our results suggest that the telomeric association of polα is independent of Stn1 (Figure 6). Together with recent report showing that polα recruitment to ss DNA is independent of xStn1 in Xenopus extract49, we conclude that CST is likely dispensable for polα recruitment to telomeres and that unidentified proteins may recruit polα to telomeres.
In yeasts and plants, homologs of CST appear to ensure telomere stability by capping chromosome ends3,11,12,13,14,15,16,17,18,19,20,21,22. In vertebrates, the capping function is achieved by shelterin proteins. Absence of shelterin components leads to telomere deprotection and prominent DDR at telomeres. The report that Stn1 associates with TPP15 has raised the speculation that hCST may also participate in telomere capping, perhaps by forming CST-shelterin subcomplexes. However, our results suggest that hStn1 plays a less important role in telomere capping. First, Stn1 unlikely limits excessive end resection at C-strand (Figure 5). Additionally, no ATM/ATR activation was observed immediately after Stn1 downregulation, but rather appeared gradually (Figure 3E). This is in striking contrast to Pot1, deficiency of which triggers pronounced telomere DNA damage and ATR activation independent of telomere length36,38,39.
Our results also reveal that the function of human Stn1 is somewhat different from the Cdc13/Stn1/Ten1 complex in budding yeast in several aspects. The budding yeast CST complex restricts excessive C-strand resection from telomere ends, interacts with polα to mediate C-strand fill-in, and regulates telomerase activity14,15,16,17,18,19,20,21,22. While hStn1 may retain the functions in telomere-specific C-strand fill-in and regulating telomerase, it may have no role in restraining excessive end resection at leading daughter telomeres (Figure 5D). It remains to be determined how end resection is controlled at human telomeres. One candidate may be hPot1, as it defines the precision of the final nucleotide at C-strand50 and mammalian Pot1b protects C-strand from excessive degradation39,51. However, depletion of human Pot1 leads to G-overhang shortening2,50, contradicting the expectation from protecting C-strand from degradation. Such paradox highlights the complexity of telomere end protection in human cells and underscores the importance to determine how CST and shelterin coordinate to protect telomeres.
Our findings reinforce the concept that long telomeric repeats may be more likely to become fragile, and have an increased chance of fork stalling during replication. Stalled forks need to be restarted in a timely manner to avoid fork collapse and telomere breakage. In the genomic region, stalled fork uncouples the replicative helicase from DNA polymerases, leading to accumulation of ss DNA. The accumulated ss DNA is then bound by RPA, which activates ATR-mediated checkpoint and stabilizes stalled forks52. It appears that homologous recombination (HR) is the primary pathway for resuming replication at stalled forks52,53. However, the telomeric region differs from the genomic region in that ATR activation and HR are normally repressed at telomeres by shelterins36,38. How stalled forks at telomeres are repaired and restarted is currently unclear. It is possible that accessory factors may have evolved and telomere-binding proteins may also have acquired additional functions to facilitate replication machinery efficiently passing through telomeres. Among them are TRF127,28, TRF226, the CST complex, Apollo26, BRCA229, BLM27, WRN30, RTEL154, and likely other factors remaining to be identified.
Materials and Methods
Cell culture and synchronization
All cells were cultured at 37 °C under 5% CO2 in DMEM supplemented with 10% fetal bovine serum or cosmic calf serum (Hyclone). Double thymidine block was used to synchronize HeLa cells as described previously4. Cells collected at different time points for analyzing DNA contents using a Beckman Coulter EPICS® XLTM flow cytometer. For replication stress and FT analysis, HeLa cells were treated with 0.2 μM aphidicolin for 17 h prior to preparation of metaphase spreads.
RNAi
Stn1 siRNA sequences targeting GAUCCUGUGUUUCUAGCCU (sequence 1) and GCUUAACCUCACAACUUAA (sequence 2) were used in previous studies4. The corresponding shRNA sequence was cloned into pSIREN-retro-puro (Clontech). Infection and selection were performed following the manufacturer's instruction. The majority of experiments were performed with sequence 2 unless specified.
Site-directed mutagenesis
QuickChange Site-directed Mutagenesis kit (Stratagene) was used to introduce mismatch mutations at siRNA targeting region in pBabe-FLAG-Stn15 to create siRNA-resistant Stn1.
Antibodies
The following primary antibodies were used: monoclonal anti-Stn1 (Santa Cruz, used in Figure 1 and then became unavailable), rabbit polyclonal anti-Stn1 (Santa Cruz, used in all other figures), monoclonal anti-actin (BD Biosciences), rabbit polyclonal anti-Pot1 (Imgenex), anti-pChk1-S345 (Cell Signaling), anti-pChk2-T68 (Cell Signaling), anti-Rad51 (Santa Cruz), anti-RPA32 (Bethyl), anti-γH2AX-S139 (Active Motif), goat polyclonal anti-polα (Santa Cruz). Secondary antibody was horseradish peroxidase-conjugated anti-mouse IgG, anti-rabbit IgG (BD Biosciences), and anti-goat IgG (Santa Cruz).
Telomeric G-overhang measurement
The mean length of the telomeric G-overhang was measured by telomere overhang protection assay as described previously41.
Non-denaturing in-gel hybridization assay
Non-denaturing in-gel hybridization was performed as described previously4.
Separation of leading and lagging daughter telomeres
Separation of leading and lagging telomeres was performed essentially as described4.
Purification of His6-Stn1 protein
Stn1 cDNA was amplified from pBabe-FLAG-Stn15 and inserted into pET22b (Clontech). Protein was induced with 0.1 mM IPTG at 37 °C for 4 h in E. coli BL21(DE3). After induction, cells were collected at 4 °C with centrifugation, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), and lysed with lysozyme (2 mg/ml) on ice for 30 min, followed by sonication on ice. Cell lysates were centrifuged at 21 000 ×g for 10 min at 4 °C to separate soluble and insoluble fractions. The majority of His6-Stn1 was in the insoluble fraction. Pellet was resuspended in resuspension buffer (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 7.9), incubated with Ni+ agarose beads (ThermoFisher) for 1 h at room temperature, and loaded on column. Non-specific bound materials were washed twice with resuspension buffer, followed by 4 times wash of buffer C (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 6.3). Bound protein was eluted with buffer D (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 5.5), followed by elution with buffer E (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 4.4).
ChIP and co-immunoprecipitation
Quantitative ChIP was performed essentially as described previously55. Co-immunoprecipitation was performed as previously described56.
Senescence-associated β-galactosidase staining
Senescence-associated β-galactosidase staining was performed for 6 h at 37 °C as described previously57.
FISH and CO-FISH
Cells were treated with colcemid, collected by trypsinization, swollen in 0.075 M KCl at 37 °C for 15 min, and fixed in a freshly prepared 3:1 mix of methanol:glacial acetic acid. Preparations were dropped onto slides and let to dry overnight. Hybridization to peptide nucleic acid (PNA) probe Alexa488-OO-(TTAGGG)3 or Cy3-OO-(CCCTAA)3 was carried out following standard FISH and CO-FISH staining58.
STELA
STELA was performed as described4. Briefly, 20 ng EcoRI digested genomic DNA was ligated to each telorette (10−3 μM) at 35 °C for 16 h in 10 μl ligation reaction containing 1× ligase buffer and 200 U T4 ligase (NEB). The ligated DNA was diluted to 250 pg/μl for subsequent PCRs. Multiple PCR reactions (30 cycles of 95 °C for 15 s, 58 °C for 20 s, and 68 °C for 10 min) were carried out in 25 μl containing 250 pg ligated DNA, 0.2 μM primers (XpYpE2 forward primer and C-teltail reverse primer), and 2 U FailSafe enzyme mix (Epicentre). DNA ligated to Tel3 (ending with the sequence CTAACC-3′) was used alone in PCR, while DNA ligated to the other five telorettes were combined together and used for PCR. Four independent PCR reactions were carried out for each sample and PCR products were resolved on 1% agarose gels in separate lanes. After electrophoresis, DNA was transferred onto positively-charged nylon membranes (Hybond N+, GE healthcare), followed by UV crosslinking and hybridization with a XpYp subtelomeric probe. Signals were detected by PhosphorImager screen (GE healthcare). For determining the contribution of internal ss G-rich DNA to short STELA products, genomic DNA was treated with or without ExoI at 37 °C for 11 h in 20 μl buffer containing 10 mM HEPES pH 7.5, 100 mM LiCl, 2.5 mM MgCl2, 5 mM CaCl2, 20 mM β-mercaptoethanol. ExoI was then heat inactivated at 70 °C for 30 min. To completely remove ExoI, samples were then treated with Proteinase K (Roche) at 55 °C for 30 min, followed by heat inactivation at 70 °C for 30 min. DNA was phenol extracted, ethanol precipitated and used in STELA as described above. Ligated DNA from all six telorettes was mixed together for PCR.
Immunofluorescence-FISH
Cells were grown in chamber slides and fixed in 4% paraformaldehyde, permeabilized with 0.15% Triton X-100, blocked with 3% BSA at 37 °C for 1 h in humidified chamber, incubated with anti-γ-H2AX for 1 h at 37 °C, washed with PBS three times, and then incubated with Dylight 549-conjugated secondary antibody (Fisher) at 37 °C for 1 h. Following γ-H2AX staining, the slides were refixed with 4% paraformaldehyde for 10 min, dehydrated in 70%, 85%, and 100% ethanol, air dried, and hybridized to Alexa488-conjugated PNA telomere probe (Panagene). The slides were denatured for 5 min at 90 °C, incubated overnight at room temperature, and washed with 70% formamide in 10 mM Tris pH 7.5 for 15 min twice, followed by three times of wash in 0.1 M Tris pH 7.5/0.15 M NaCl/0.08% Tween-20 for 5 min each. Slides were then dehydrated in ethanol series, DNA was counterstained with DAPI, and Z-stack images were taken at a 0.275 μM thickness per slice under Zeiss AxioImager M2 epifluorescence microscope.
References
Palm W, de Lange T . How shelterin protects mammalian telomeres. Annu Rev Genet 2008; 42:301–334.
Miyake Y, Nakamura M, Nabetani A, et al. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 2009; 36:193–206.
Surovtseva YV, Churikov D, Boltz KA, et al. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 2009; 36:207–218.
Dai X, Huang C, Bhusari A, Sampathi S, Schubert K, Chai W . Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J 2010; 29:2788–2801.
Wan M, Qin J, Songyang Z, Liu D . OB-fold containing protein 1 (OBFC1), a human homologue of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 2009; 284:26725–26731.
Casteel DE, Zhuang S, Zeng Y, et al. A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 2009; 284:5807–5818.
Levy D, Neuhausen SL, Hunt SC, et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc Natl Acad Sci USA 2010; 107:9293–9298.
Anderson BH, Kasher PR, Mayer J, et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 2012; 44:338–342.
Polvi A, Linnankivi T, Kivela T, et al. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet 2012; 90:540–549.
Keller RB, Gagne KE, Usmani GN, et al. CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr Blood Cancer 2012; 59:311–314.
Martin V, Du LL, Rozenzhak S, Russell P . Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA 2007; 104:14038–14043.
Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE . STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA 2008; 105:19815–19820.
Grandin N, Damon C, Charbonneau M . Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J 2001; 20:6127–6139.
Grandin N, Damon C, Charbonneau M . Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J 2001; 20:1173–1183.
Qi H, Zakian VA . The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev 2000; 14:1777–1788.
Chandra A, Hughes TR, Nugent CI, Lundblad V . Cdc13 both positively and negatively regulates telomere replication. Genes Dev 2001; 15:404–414.
Grandin N, Damon C, Charbonneau M . Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol 2000; 20:8397–8408.
Grandin N, Reed SI, Charbonneau M . Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev 1997; 11:512–527.
Nugent CI, Hughes TR, Lue NF, Lundblad V . Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 1996; 274:249–252.
Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI . Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol 2006; 8:748–755.
Petreaca RC, Chiu HC, Nugent CI . The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics 2007; 177:1459–1474.
Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D . Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J 2008; 27:2328–2339.
Gilson E, Geli V . How telomeres are replicated. Nat Rev Mol Cell Biol 2007; 8:825–838.
Sampathi S, Chai W . Telomere replication: poised but puzzling. J Cell Mol Med 2011; 15:3–13.
Miller KM, Rog O, Cooper JP . Semi-conservative DNA replication through telomeres requires Taz1. Nature 2006; 440:824–828.
Ye J, Lenain C, Bauwens S, et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 2010; 142:230–242.
Sfeir A, Kosiyatrakul ST, Hockemeyer D, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009; 138:90–103.
Martinez P, Thanasoula M, Munoz P, et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 2009; 23:2060–2075.
Badie S, Escandell JM, Bouwman P, et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 2010; 17:1461–1469.
Crabbe L, Verdun RE, Haggblom CI, Karlseder J . Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 2004; 306:1951–1953.
Chai W, Du Q, Shay JW, Wright WE . Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 2006; 21:427–435.
Saharia A, Teasley DC, Duxin JP, Dao B, Chiappinelli KB, Stewart SA . FEN1 ensures telomere stability by facilitating replication fork re-initiation. J Biol Chem 2010; 285:27057–27066.
Zou L, Elledge SJ . Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542–1548.
Zou L . Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev 2007; 21:879–885.
Guo X, Deng Y, Lin Y, et al. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J 2007; 26:4709–4719.
Denchi EL, de Lange T . Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007; 448:1068–1071.
Flynn RL, Centore RC, O'Sullivan RJ, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011; 471:532–536.
Wu L, Multani AS, He H, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 2006; 126:49–62.
He H, Wang Y, Guo X, et al. Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol 2009; 29:229–240.
Zhao Y, Sfeir AJ, Zou Y, et al. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 2009; 138:463–475.
Chai W, Shay JW, Wright WE . Human telomeres maintain their overhang length at senescence. Mol Cell Biol 2005; 25:2158–2168.
Goulian M, Heard CJ, Grimm SL . Purification and properties of an accessory protein for DNA polymerase alpha/primase. J Biol Chem 1990; 265:13221–13230.
Goulian M, Heard CJ . The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J Biol Chem 1990; 265:13231–13239.
Gu P, Min JN, Wang Y, et al. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J 2012; 31:2309–2321.
Zhu W, Ukomadu C, Jha S, et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev 2007; 21:2288–2299.
Chattopadhyay S, Bielinsky AK . Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell 2007; 18:4085–4095.
Ricke RM, Bielinsky AK . Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol Cell 2004; 16:173–185.
Verdun RE, Karlseder J . The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 2006; 127:709–720.
Nakaoka H, Nishiyama A, Saito M, Ishikawa F . Xenopus laevis Ctc1-Stn1-Ten1 (xCST) complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 2012; 287:619–627.
Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T . POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 2005; 24:2667–2678.
Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T . Engineered telomere degradation models dyskeratosis congenita. Genes Dev 2008; 22:1773–1785.
Petermann E, Helleday T . Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 2010; 11:683–687.
Allen C, Ashley AK, Hromas R, Nickoloff JA . More forks on the road to replication stress recovery. J Mol Cell Biol 2011; 3:4–12.
Vannier J-BP-K, Visnja, Petalcorin MIR, Hao D, Boulton SJ . RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012; 149:795–806.
Dai X, Huang C, Chai W . CDK1 differentially regulates G-overhang generation at leading- and lagging- strand telomeres in telomerase negative cells in the G2 phase. Cell Cycle 2012; 11:3079–3086.
Sampathi S, Bhusari A, Shen B, Chai W . Human Flap Endonuclease I is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J Biol Chem 2009; 284:3682–3690.
Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92:9363–9367.
Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH . Strand-specific postreplicative processing of mammalian telomeres. Science 2001; 293:2462–2465.
Acknowledgements
We thank S Chang for communicating results prior to publication. We also thank Z Songyang (Baylor College of Medicine, USA) for pBabe-FLAG-Stn1. This work was supported by NIH R15GM099008 and American Cancer Society Institutional Research Grant IRG-77-003-32 to WC.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Anti-Stn1 antibody recognizes Stn1 protein. (PDF 417 kb)
Supplementary information, Figure S2
No increase in T-SCE or signal free ends (SFE) after Stn1 depletion in HeLa 1.2.11. (PDF 212 kb)
Supplementary information, Figure S3
Separation of replicated and unreplicated telomeres using CsCl ultracentrifugation. (PDF 934 kb)
Supplementary information, Figure S4
Growth curve of BJ and BJ/E6/E7 expressing shLuc and shStn1. (PDF 193 kb)
Supplementary information, Figure S5
Short STELA products resulted largely from telorettes ligating at telomere ends. (PDF 586 kb)
Supplementary information, Figure S6
Co-IP of polα and Stn1 throughout the cell cycle. (PDF 293 kb)
Rights and permissions
About this article
Cite this article
Huang, C., Dai, X. & Chai, W. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res 22, 1681–1695 (2012). https://doi.org/10.1038/cr.2012.132
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2012.132
Keywords
This article is cited by
-
TNF-α impairs EP4 signaling through the association of TRAF2-GRK2 in primary fibroblast-like synoviocytes
Acta Pharmacologica Sinica (2022)
-
Crosstalk between CST and RPA regulates RAD51 activity during replication stress
Nature Communications (2021)
-
Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization
Nature Reviews Molecular Cell Biology (2021)
-
Telomere Maintenance Genes are associated with Type 2 Diabetes Susceptibility in Northwest Indian Population Group
Scientific Reports (2020)
-
Tpz1TPP1 prevents telomerase activation and protects telomeres by modulating the Stn1-Ten1 complex in fission yeast
Communications Biology (2019)