Abstract
MYB-type transcription factors contain the conserved MYB DNA-binding domain of approximately 50 amino acids and are involved in the regulation of many aspects of plant growth, development, metabolism and stress responses. From soybean plants, we identified 156 GmMYB genes using our previously obtained 206 MYB unigenes, and 48 were found to have full-length open-reading frames. Expressions of all these identified genes were examined, and we found that expressions of 43 genes were changed upon treatment with ABA, salt, drought and/or cold stress. Three GmMYB genes, GmMYB76, GmMYB92 and GmMYB177, were chosen for further analysis. Using the yeast assay system, GmMYB76 and GmMYB92 were found to have transactivation activity and can form homodimers. GmMYB177 did not appear to have transactivation activity but can form heterodimers with GmMYB76. Yeast one-hybrid assay revealed that all the three GmMYBs could bind to cis-elements TAT AAC GGT TTT TT and CCG GAA AAA AGG AT, but with different affinity, and GmMYB92 could also bind to TCT CAC CTA CC. The transgenic Arabidopsis plants overexpressing GmMYB76 or GmMYB177 showed better performance than the GmMYB92-transgenic plants in salt and freezing tolerance. However, these transgenic plants exhibited reduced sensitivity to ABA treatment at germination stage in comparison with the wild-type plants. The three GmMYB genes differentially affected a subset of stress-responsive genes in addition to their regulation of a common subset of stress-responsive genes. These results indicate that the three GmMYB genes may play differential roles in stress tolerance, possibly through regulation of stress-responsive genes.
Similar content being viewed by others
Introduction
Since the first MYB gene, the oncogene v-myb, was identified from the avian myeloblastosis virus 1, many MYB genes have been found in nearly all eukaryotes. MYB proteins can be classified into three subfamilies depending on the numbers (one, two or three) of adjacent repeats in the MYB domain 2, 3, 4, and MYB genes encoding two repeats (i.e. R2R3-MYB) constitute the largest MYB subfamily in plants 5, 6. Members of the MYB family have been found to be involved in diverse processes 7, including developmental control and determination of cell fate and identity 8, 9, plant responses to environmental factors and hormones 3, 10, 11, 12, signal transduction in plant growth processes 13, 14, pathogen defense 15, and xylogenesis and lignin biosynthesis 16, 17, 18, 19, 20, 21.
When plants are exposed to extreme conditions such as high salt, cold, and drought, plant growth and development are delayed. Analysis of gene expressions in Arabidopsis has revealed that a number of genes including effector genes and regulatory genes are involved in responses to various stresses. These genes belong to several independent signaling pathways 22, 23. Because multiple stress responses are necessary for plants to endure severe stress conditions, the engineering of a single enzyme may not be sufficient 24, 25. However, it is possible for a single transcription factor to control the expression of multiple target genes through specific binding of the transcription factor to a cis-acting element in the promoters of their respective target genes 26. Promoter analyses of these stress-inducible genes have identified at least four independent regulatory systems for gene expression 27, 28, 29. There are two major cis-acting elements: ABRE and DRE/CRT. ABRE is involved in ABA-dependent gene expression in osmotic and cold stress responses, and DRE/CRT is involved in ABA-independent gene expression in osmotic and cold stress responses 30. However, these two regulatory systems of gene expression are interdependent in the stress-responsive expression of the rd29A gene 31.
Soybean (Glycine max) is the major source of edible vegetable oil, and the dominant source of high-quality protein for livestock and humans. Its growth and yield were affected by various abiotic stresses. Several abiotic stress-inducible genes have been identified from soybean plants and their physiological functions have been characterized 32, 33, 34, 35, 36, 37, 38. However, few soybean MYB transcription factor genes related to abiotic stresses were reported. In the present study, the expressions of soybean MYB transcription factor genes were analyzed in response to drought, salt, and cold treatments. Three stress-inducible genes, GmMYB76, GmMYB92, and GmMYB177, were further characterized for their roles in stress tolerance. We find that the transgenic Arabidopsis plants overexpressing GmMYB76, GmMYB92, or GmMYB177 show reduced sensitivity to ABA and exhibit better tolerance to salt and freezing treatments when compared with the wild-type (WT) plants.
Results
Identification and phylogenetic analysis of MYB genes from soybean
Our previous study has identified 206 MYB genes from 56 147 soybean unigenes 39. After further analysis by blasting these sequences against those from Genbank and through comparison of the amino acid sequences, 156 MYB genes were confirmed in the present study. These genes were named as GmMYB17-GmMYB187 because the GmMYB1-GmMYB16 have been used by others for MYB-related expressed sequence tags (ESTs) (Supplementary information, Table S1). In our nomenclature, some genes are not named in sequential numbers due to the removal of sequences of low quality. Twelve additional MYB genes from soybean plants, including eight genes with full-length open reading frames, have been given other names (Supplementary information, Table S1). In our analysis, 48 genes with full-length open reading frames were identified. Totally 184 MYB genes from soybean are listed in Supplementary information, Table S1.
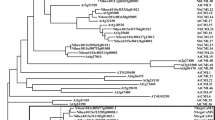
The evolutionary relationships among 36 MYB proteins from Arabidopsis and 55 full-length MYB proteins available from soybean were analyzed using their full-length amino acid sequences (Figure 1). The 36 MYB proteins from Arabidopsis are selected from different subgroups described previously 4, 40. Figure 1 shows that most of the GmMYB proteins clustered with the Arabidopsis members, which belong to different categories. However, a few GmMYB members cannot be classified into any categories due to absence of conserved motifs.
Cluster analysis of 55 soybean full-length GmMYB proteins with 36 Arabidopsis MYB proteins. The 36 MYB proteins from Arabidopsis are selected from different subgroups described previously, and the consensus sequences of each group were also shown based on the analysis of Arabidopsis MYB proteins 4, 40.
Expression of GmMYB genes under various stresses
Expression of the GmMYB genes in response to abiotic stresses was investigated using RT-PCR. Among 156 GmMYBs, the expression of 43 members responded to at least one of the four treatments including salt, cold, drought, and ABA (Supplementary information, Table S1). Expression of six genes was altered upon all four treatments. Ten genes were responsive to all the three abiotic stresses (Figure 2A). The overall expression of 26.3% of the GmMYB genes was changed upon treatment with at least one of the three abiotic stresses.
Expressions of GmMYB76, GmMYB92, and GmMYB177 in soybean plants. (A) Comparison of numbers of soybean GmMYB genes that are responsive to salt, drought, and cold treatments. (B) Expression of GmMYB76, GmMYB92, and GmMYB177 in different soybean organs. (C) Expression of GmMYB76, GmMYB92, and GmMYB177 in response to ABA, cold, drought, and salt treatments. Water treatment was performed as a control treatment. Tubulin gene was amplified as a control.
Three GmMYB genes GmMyb76, GmMYB92, and GmMYB177, which were responsive to abiotic stresses, were cloned and further analyzed. GmMYB76 and GmMYB92 belong to group 20 and 7, respectively, whereas the GmMYB177 belongs to one MYB domain CCA1-like type (Figure 1). The expressions of the three GmMYBs were monitored in cotyledons, roots, stems, leaves, and flowers of soybean plants. As shown in Figure 2B, the expression of GmMYB76 was higher in cotyledons, roots, stems, and flowers than in leaves. The expression of GmMYB92 was high in cotyledons and roots but very low in stems, flowers, or leaves. The expression of GmMYB177 was higher in cotyledons and flowers than in stems, roots, or leaves.
The expressions of the three GmMYB genes under various treatments were also investigated. Figure 2C showed that GmMYB76 was induced by NaCl treatment but not by other treatments. GmMYB92 was induced by both cold and salt treatments, whereas GmMYB177 was induced by drought and NaCl treatments. It should be noted that none of the three genes were affected by ABA treatment.
Analysis of transactivation and dimerization abilities of the three GmMYBs
The transactivation abilities of GmMYB76, GmMYB92, and GmMYB177 were analyzed using a yeast assay system. GAL4 DNA binding domain (BD)-GmMYBs fusion plasmids were transformed into yeast cells and assayed for their ability to activate transcription of the marker HIS3 gene controlled by the GAL4 upstream activation sequence and their ability to promote yeast growth in the absence of histidine. β-Galactosidase activity was also examined for the marker LacZ gene expression. Figure 3A showed that full-length GmMYB76 and GmMYB92 had transactivation capacity whereas GmMYB177 did not.
Transcriptional activation, dimerization, and DNA-binding analysis of GmMYB76, GmMYB92, and GmMYB177. (A) Transactivation ability of GmMYB76, GmMYB92, and GmMYB177. Growth of the transformants on SD/−His and blue color in β-gal assay indicate that the corresponding gene has transactivation activity. (B) Dimerization analysis among the three GmMYBs or between the three GmMYBs and GmbZIP46. Only the positive interactions were shown. The CK+(WT) containing pAD-WT/pBD-WT pair indicates positive interaction control and the pAD-GmMYBs/pBD or pAD-GmbZIP46/pBD pairs indicates negative controls. Growth of the transformants on SD(−Trp−Leu−His plus 3-AT) and the blue color in β-gal assay indicate that the corresponding protein pairs have the ability to form dimers. (C) DNA-binding ability of the GmMYB76, GmMYB92, and GmMYB177 by using yeast one-hybrid system. CK− indicates growth of cells harboring the effector plasmids pAD-GmMYBs and the reporter plasmid pHIS2 (without cis-DNA elements). Growth of the transformants on SD(−Trp−Leu−His plus 3-AT) indicates that the GmMYB proteins can bind to the corresponding cis-DNA elements. MRE1 (CCG GCA GTT AGG AT), mMRE1 (CCG GAA AAA AGG AT); MBSI (TAT AAC GGT TTT TTA), mMBSI (TAT AAA AAT TTT TTT A); MRE3 (TCT AAC CTA CCA), mMRE3 (TCT AAA AAA CCA); MRE4 (TCT CAC CTA CC), mMRE4 (TCT CGA TCA CC).
MYB proteins can form homo- and/or heterodimers to exert their physical and biological functions, as in the case of RTBP1 (rice telomere-binding protein1), which can form homodimers to bind telomeric DNA 41. In order to analyze the dimerization abilities between these three GmMYB proteins, the yeast two-hybrid system was employed using two different reporters, lacZ and HIS3, in YRG-2 yeast cells. The full-length GmMYB proteins were expressed as fusions to the yeast GAL4 activation domain (AD) and GAL4 DNA-BD. The yeast strain was co-transformed with the pAD-GmMYB and the pBD-GmMYB. Then the transformants and controls were selected on SD/−Trp−Leu−His+10 mM 3-AT and examined for β-galactosidase activity. Figure 3B showed that GmMYB76 can form a homodimer whereas GmMYB76 and GmMYB177 can form a heterodimer. Because cells harboring pBD-GmMYB92 plus pAD vector can grow on SD/−Trp−Leu−His+10 mM 3-AT (data not shown) and this will generate false positive response, we removed the C-terminal portion of the GmMYB92 to make a pBD-GmMYB92-N. The cells harboring pBD-GmMYB92-N or pBD-GmMYB92-N plus pAD vector could not grow on SD(−His) or SD/−Trp−Leu−His+10 mM 3-AT (Figure 3A; data not shown), and thus pBD-GmMYB92-N can be used for examination of dimerization with other proteins. Figure 3B showed that cells harboring pBD-GmMYB92-N plus the pAD-GmMYB92 can grow on selection medium, suggesting that GmMYB92 can form a homodimer and the N-terminal portion is responsible for the dimerization. GmMYB92-N cannot form dimers with other proteins (data not shown).
MYB proteins can interact with other transcriptional regulators 42, 43, 44. We analyzed the interaction between these GmMYBs and five GmbZIP proteins, whose genes were identified from soybean and were responsive to abiotic stresses (Liao Y, Zhang JS and Chen SY, unpublished results). We observed an interaction between GmMYB76 and GmbZIP46 (Figure 3B). There were no interactions between GmMYBs and other GmbZIPs (data not shown).
DNA-binding ability of the three GmMYBs
The cis-DNA elements for several MYB proteins have been identified 45. We investigated if GmMYB76, GmMYB92, and GmMYB177 can bind to these elements in yeast one-hybrid assay system. Four cis-DNA elements MRE1 (CCG GCA GTT AGG AT), MBSI (TAT AAC GGT TTT TTA), MRE3 (TCT AAC CTA CCA) and MRE4 (TCT CAC CTA CC) and their corresponding mutant sequences mMRE1(CCG GAA AAA AGG AT), mMBSI (TAT AAA AAT TTT TTT A), mMRE3 (TCT AAA AAA CCA), and mMRE4 (TCT CGA TCA CC) were synthesized and inserted into the reporter plasmid pHIS2 containing the reporter gene HIS3 46, 47. A minimal promoter was present downstream of the cis-elements but upstream of the HIS3 gene. Each of the reporter plasmid harboring different elements plus the reporter gene HIS3, together with the effector plasmid pAD-GmMYB76, 92 and 177, was transformed into yeast (Y187) cells. The transformants and controls were selected on SD/−Trp−Leu−His plus 3-AT, and growth of the transformants indicates binding of the protein to the corresponding cis-DNA elements. Figure 3C showed that all the three proteins GmMYB76, GmMYB92, and GmMYB177 could bind to the sequence MBSI (TAT AACGGT TTT TT). GmMYB92 could also bind to the MRE4 sequence (TCT CACCTA CC) and mMRE1 (CCG GAAAAAAGG AT). GmMYB76 and GmMYB177 bound to the element mMRE1 with a weak affinity. None of the proteins could bind to MRE1, mMBSI, mMRE3, MRE3, or mMRE4. These results indicate that the three proteins can differentially bind to the cis-DNA elements examined.
ABA sensitivity of the transgenic Arabidopsis plants overexpressing the three GmMYBs
To investigate the in vivo function in plants, the GmMYB76, GmMYB92, and GmMYB177 genes were inserted in sense orientation into a binary vector pBIN438 in which the GmMYBs were under the control of the 2× CaMV 35S promoter. The genes were transformed into Arabidopsis by using the vacuum infiltration method. The homozygous T3 lines, with relatively higher expression of GmMYB76, GmMYB92, or GmMYB177, were used for further analysis (Figure 4A). No obvious differences were observed in the phenotypes between the transgenic plans harboring GmMYB76, GmMYB92, or GmMYB177 and the WT plants grown on MS agar plates and in the soil (Supplementary information, Figure S1; data not shown).
ABA sensitivity of the transgenic plants overexpressing the GmMYB76, GmMYB92, and GmMYB177. (A) Expression of GmMYB76, GmMYB92, and GmMYB177 in various transgenic lines as revealed by RT-PCR. Actin gene was amplified as a control. (B) Germination of the GmMYB-transgenic seeds under various ABA concentrations. Each data point represents the mean of triplicates and each measurement has more than 100 seeds. The bars indicate SD. * indicates that the differences between the transgenic line and the Col-0 are significant (P<0.05). ** indicate that the differences between the transgenic line and the Col are highly significant (P<0.01).
To investigate whether overexpression of GmMYB76, GmMYB92, and GmMYB177 affects ABA sensitivity of the transgenic plants, the seeds from the transgenic plants and WT plants were germinated in the medium containing ABA and the germination rates were compared. Figure 4B showed that, at 0.75 μM or higher concentrations of ABA, the germination rates of all the transgenic lines were substantially higher than those of the WT plants. At lower ABA concentrations, the germination rates were not significantly affected in transgenic seeds. These results indicate that the transgenic seeds were less sensitive to ABA treatment.
Performance of the GmMYB-transgenic plants under salt stress
Because the expression of GmMYB76, GmMYB92, and GmMYB177 was induced under salt stress, these genes may participate in salt tolerance in plants. The GmMYB-transgenic seeds were germinated on NaCl medium and their germination rates were compared. Figure 5A showed that, at 100 mM NaCl, the germination rates of the GmMYB177-transgenic plants were higher than those of WT plants and other transgenic plants. At 150 mM NaCl no WT seeds could germinate, whereas around 10%, 5%, and 15% of the GmMYB76-, GmMYB92-, and GmMYB177-transgenic seeds, respectively, could germinate. The plate-grown seedlings were also transferred to soil and treated with stepwise increasing concentrations of NaCl. Figure 5B and 5C showed that, with this treatment, the GmMYB76- and GmMYB177-trangenic plants grew better and higher than WT plants. However, the growth of GmMYB92-transgenic plants showed no significant difference in comparison with WT plants under the stress. In the absence of stress, all the transgenic plants showed no significant difference compared to WT plants (Supplementary information, Figure S1). The plate-grown seedlings were also transferred to soil and further treated with high salt (600 mM NaCl) for 12 days. The plant survival was scored and compared. Figure 5D and 5E showed that the survival rates of the GmMYB76-, GmMYB92-, and GmMYB177-transgenic plants were significantly higher than those of WT plants. All these results indicate that the transgenic plants show better tolerance to salt stress than WT plants. However, each of the three GmMYB genes may confer differential tolerance to salt stress at different developmental stages and with different treatments.
Effects of salt stress on the transgenic plants overexpressing the GmMYB76, Gm92, and GmMYB177. Transgenic lines 76–23.5, 76-31, or 76-45 for GmMYB76, 92-30 and 92-33 for GmMYB92, and 177-32, 177-35, 177-36 for GmMYB177 were used. (A) Comparison of the germination rates of the transgenic seeds under salt stress. Experiments were performed in triplicate and bars indicate SD. (B) Phenotypic comparison of the plant growth at bolting stage under salt stress. Two-week-old seedlings were transferred to soil for one week, and then treated with increasing concentrations of NaCl. Photographs were taken after 14 days treatment. The plant height was measured after treatment for 20 days. (C) Effect of salt stress on the inflorescence height of the plant grown in soil. Plant height in (B) was measured. Each data point represents the mean of triplicates and each measurement has 11 plants. Bars indicate SD. (D) Survival of the transgenic plants under high salt (600 mM NaCl) treatment for 12 days. (E) Comparison of the survival rate of the transgenic plants in (D). Each data point represents the mean of triplicate tests and each test has 18 plants. Bars indicate SD. For (C and E), * indicates that the differences between the transgenic line and the Col-0 are significant (P<0.05). ** indicate that the differences between the transgenic line and the Col-0 are highly significant (P<0.01).
Performance of the GmMYB-transgenic plants under freezing stress
Because the expression of GmMYB92 was induced by cold treatment in soybean plants, we examined the effect of freezing on the performance of the GmMYB-transgenic plants. Soil-grown seedlings were subjected to treatment at −6 °C for 2 d, and the survival was evaluated after recovery for 3 days under normal conditions. The line 76–23.5 for the GmMYB76, and the lines 177-32 and 177-35 for the GmMYB177 had a survival percentage significantly higher than that of WT plants (Figure 6A and 6B). Other lines showed no significant difference or only slightly higher survival percentage compared to WT. These results imply that the GmMYB177 gene may confer freezing tolerance in the transgenic Arabidopsis plants whereas GmMYB76 may play partial roles in freezing tolerance.
Performance of the GmMYB-transgenic plants upon freezing and comparison of proline contents. Transgenic lines 76–23.5, 76-31, and 76-45 for GmMYB76, 92-30 and 92-33 for GmMYB92, and 177-32, 177-35, and 177-36 for GmMYB177 were used for the analysis. (A) Phenotype of the GmMYB-transgenic plants after freezing treatment. Two-week-old seedlings were transferred to soil for one week, and treated at −6 °C for 2 days. The photograph was taken after recovery for 7 days, and survival rate was also calculated. (B) Comparison of the survival of the transgenic plants in (A). Each data point represents the mean of triplicate tests and each test has 18 plants. Bars indicate SD. (C) Proline content in the GmMYB-transgenic lines. Each data point represents the mean of three replicates and bars indicate SD. For (B and C), * indicates that the differences between the transgenic line and the Col are significant (P<0.05). ** indicate that the differences between the transgenic line and the Col are highly significant (P<0.01).
Proline is one of the osmolytes that copes with cellular dehydration and, in several plant species, the increase in proline content during cold correlates with freezing tolerance 48, 49. The proline levels were measured in 16-day-old WT plants and the transgenic plants grown under normal condition. Figure 6C showed that the level of free proline was significantly higher in all the GmMYB-transgenic plants than in the WT plants. The higher proline levels may contribute to the freezing tolerance of the GmMYB-transgenic plants.
Altered gene expression in the GmMYB-transgenic plants
Because overexpression of GmMYB76, 92, and 177 led to stress tolerance and changed ABA response in transgenic Arabidopsis plants, we examined if abiotic stress- and ABA-responsive genes were altered in these plants. As shown in Figure 7, the transcript levels of a number of ABA- or abiotic stress-responsive genes were changed between the transgenic plants and WT plants. The expression levels of rd29B 50, DREB2A 51, 52, P5CS 53, 54, RD17 55, ERD10 55, 56, and COR78/rd29A 57, 58 were enhanced in the GmMYB76-transgenic plants. The expressions of DREB2A, RD17, and P5CS were higher in GmMYB92-transgenic plants than in WT plants. However, the expressions of rd29B, COR6.6, COR15a 59, and COR78/rd29A were lower in these GmMYB92-transgenic plants. In the GmMYB177-transgenic plants, genes including rd29B, ABI2 60, DREB2A, RD17, P5CS, ERD10, COR6.6 55, 61, ERD11 62, and COR78 were upregulated. The expressions of rab18 63 and ABI1, which were induced by ABA, were not significantly affected in the three GmMYB-transgenic plants when compared with the WT plants. These results suggest that each of the three GmMYB genes may differentially regulate the expressions of the downstream genes, which are related to abiotic stresses and/or ABA responses.
Expression of ABA- and abiotic stress-responsive genes in GmMYB-transgenic plants. Total RNAs were isolated from two-week-old plants grown on MS plates and then subjected to RT-PCR analysis. Transgenic lines 76-31 and 76-45 for GmMYB76, 92-30 and 92-33 for GmMYB92, and 177-32, 177-35, and 177-36 for GmMYB177 were used for the examination.
Discussion
The MYB family comprises a large group of transcription factors involved in plant development and tissue differentiation, and are found in nearly all eukaryotes. They contain a DNA-binding MYB domain in the N-terminal part of the protein and bind DNA in a sequence-specific manner. MYB proteins are classified into three sub-families, namely MYB1R, MYBR2R3, and MYB3R, and most plant MYB proteins are the R2R3 type 4. The R2R3-type MYB factors encoded by the AtMYB genes from Arabidopsis have been categorized into 22 subgroups on the basis of conserved amino-acid sequence motifs present carboxyterminal to the MYB domain 40, 4. There are 203 MYB members in Arabidopsis and 206 MYB members in soybean 39. Further analysis confirmed 156 MYB members of soybean, and 55 full-length MYB proteins of soybean clustered, with some but not all the subgroups of MYB proteins from Arabidopsis. Among the 156 GmMYBs, 27% were regulated by at least one of the four treatments including ABA, salt, drought, and low temperature. Three genes, GmMYB76, GmMYB92, and GmMYB177, which were induced by abiotic stresses, were chosen for further investigation.
Most MYB proteins are presumed to be transcriptional activators with ADs in the C-terminal region to the DNA-BD since c-MYB has an acidic AD C-terminal to its DNA-BD 64. However, the sequences in the C-terminal regions of MYB proteins are not strongly conserved, presumably because the structural determinants for ADs are fairly flexible. Not all MYB-related proteins need to be transcriptional activators. A survey of the predicted C-terminal sequences of the Arabidopsis R2R3 MYB family revealed 22 different subgroups, and these subgroups showed limited sequence conservation within their C-terminal regions 40. The moderately conserved motifs in each subgroup might represent ADs, an idea supported by the fact that some are relatively acidic, and others are rich in amino acids (glutamine and proline) frequently associated with ADs. Alternatively, these regions with sequence conservation may represent repression domains or domains for interaction with other transcription factors 42. We show that GmMYB76 and GmMYB92 have transactivation activities in yeast assay system, whereas GmMYB177 has no transactivation ability. These observations need to be examined further in plant system. Both GmMYB76 and GmMYB92 can form homodimers, and GmMYB76 can form heterodimers with GmMYB177 and GmbZIP46, respectively. However, the biological function of these interactions needs to be further investigated.
Sequence-specific DNA-binding has been demonstrated for several MYB proteins, in agreement with their regulation in transcription, and the proteins can bind to one or more cis-DNA elements. Different target recognition sites for different groups of MYB proteins have been identified. Mammalian repeat MYBs such as c-MYB, A-MYB, and B-MYB and closely related proteins from invertebrates and cellular slime moulds all bind to the cognate site T/CAACG/TGA/C/TA/C/T (MBSI). Some of the two repeat R2R3-type proteins in plants can recognize this binding site while others cannot. Several plant MYB proteins that bind to MBSI will also bind to a second site, TAACTAAC (MBSII), which is a cis-DNA element recognized by the majority of plant R2R3 MYB proteins 5. GmMYB76, GmMYB92, and GmMYB177 all could bind to the sequence MBSI (TAT AACGGT TTT TT), which is the recognition site of AtMYB2 42, 10. GmMYB92 can also bind to the cis-DNA sequences MRE4 (TCT CACCTA CC) 47 and mMREI (CCG GAAAAAAGG AT), while GmMYB76 and GmMYB177 can only interact with mMREI weakly. The mMREI (CCG GAAAAAAGG AT) is similar to aaaAaaCG/CGTTA, which is bound by MmMYB, PhMYB3, and AtMYB77 5. The differential binding ability suggests that the three GmMYBs may regulate different sets of downstream genes in addition to the common sets of genes. This speculation appears to be consistent with the analysis of altered gene expressions in the GmMYB-transgenic plants (Figure 7). Whereas all the three GmMYB genes promoted the expression of DREB2A, RD17, and P5CS genes, the regulation of rd29B, ERD10, and COR78 seemed different in the GmMYB-transgenic plants. GmMYB76 and GmMYB177 can enhance the expression of rd29B, ERD10, and COR78 genes. On the contrary, GmMYB92 inhibited rd29B or COR78 but did not affect the ERD10 gene.
Functional analyses of plant MYBs indicate that they regulate numerous processes including responses to environmental stress 40, 65, 66. MYBs have been found to be involved in ABA-dependent or ABA-independent signaling pathways. The genes AtMYB2 10, CpMYB10 67, and HOS10 68 are induced by ABA, and also induced by drought, salt, chilling, or heat treatments. However, the genes HPPBF-1 69, Osmyb4 49, and AtMYB61 70 are not induced by ABA, but can enhance drought, salt, or freezing tolerances. AtMYB60 is repressed by ABA and desiccation, and a null mutation in AtMYB60 leads to reduction in stomatal opening and water stress tolerance 71. In the present study, the three MYB transcription factor genes, GmMYB76, GmMYB92, and GmMYB177, from the soybean can be induced by various abiotic stresses but are not significantly affected by ABA treatment. However, the transgenic Arabidopsis plants overexpressing these three genes exhibited reduced ABA sensitivity, suggesting that the three GmMYB genes could function as negative regulators of ABA signaling.
Studies have shown that overexpression of MYB transcription factor genes improves stress tolerance. Overexpression of CpMYB10 in Arabidopsis can enhance osmotic stress tolerance of transgenic seedlings germinated in plates, and improve desiccation and salt tolerance in adult plants grown in soil 67. Overexpression of Osmyb4 in Arabidopsis increases plant tolerance to chilling and freezing 49. Several MYB proteins have relatively high similarity (45-84%) with GmMYB76, GmMYB92, and GmMYB177. AtMYB2, AtMYB112, and a homeodomain-related protein from Medicago truncatula (ABE78836) share similarity with GmMYB76. AtMYB12, AtMYB3/4/5, and MYB56 from Vitis Vinifera (AAX51291) show similarity with GmMYB92. A homeodomain-related protein from Medicago truncatula (ABE83274) is homologous to GmMYB177. Among these homologous genes, only AtMYB2 is induced by dehydration, high salt stress, and exogenous ABA. However, no stress tolerance was reported in transgenic plants overexpressing this gene 10. Coexpression of both AtMYB2 and AtMYC2 conferred moderate stress tolerance 72. Overexpression of GmMYB76, a close homologue of AtMYB2, in Arabidopsis plants improved salt tolerance and partial freezing tolerance in the transgenic plants, implying that GmMYB76 may have a stronger role than the AtMYB2 in regulation of stress tolerance. GmMYB92 and GmMYB177 also showed dominant roles in conferring plant salt and freezing tolerance. These analyses support that multiple MYB proteins are functional in plant responses to various stresses.
The GmMYB76, GmMYB92, and GmMYB177 genes may confer stress tolerance through upregulation of downstream genes DREB2A, RD17, and P5CS. DREB2A, an AP2 domain transcription factor, is also induced by dehydration and salt stress. Overexpression of DREB2A induced weak expression of the target genes under unstressed conditions 51. However, its constitutive active form caused significance stress tolerance in transgenic plants 73. RD17 contains the DRE or related motif in its promoter region and is induced by dehydration, salt, and cold. Its overexpression increased the freezing tolerance of transgenic plants 74. DREB2A can promote the expression of RD17 73. P5CS encoding delta-pyrroline-5-C synthase was upregulated in 35S-GmMYBs transgenic plants, leading to proline accumulation (Figure 6C). Free proline was increased in plants in response to many stresses 75, and its accumulation could contribute to the increase of freezing tolerance 76 and salt tolerance 77. Therefore, GmMYB76, GmMYB92, and GmMYB177 may contribute to the basal salt tolerance at least via activation of the above three genes. However, it should be noted that the levels of DREB2A, RD17, and P5CS transcripts are much higher in the GmMYB177-transgenic plants than those in the GmMYB76- and GmMYB92-transgenic plants, consistent with the good performance of the GmMYB177-transgenic plants in germination test and freezing treatment (Figures 5A, 6A and 6B). In addition to the common set of genes, GmMYB76 and GmMYB177 also promoted expressions of a subset of genes including rd29B, ERD10, and COR78, which are involved in stress tolerance. The enhanced expression of the three genes was not found in the GmMYB92-transgenic plants. These genes may potentially contribute to the better performance of the GmMYB76- and GmMYB177-transgenic plants during germination test (Figure 5A), stepwise salt treatment (Figure 5B and 5C), and freezing treatment (Figure 6A and 6B). The differential expression of the downstream genes may depend on the differential DNA-binding ability of the three GmMYBs (Figure 3C), which is determined by the MYB domain structures. It should be noted that, although GmMYB177 and GmMYB76 regulated similar genes, their structures are different, with GmMYB177 being the 1R-type MYB protein and GmMYB76 being the R2R3-type MYB protein. It is possible that the MYB repeat in GmMYB177 is functionally equivalent to the MYB repeat in GmMYB76 in regulation of gene expressions. The relationship between structure and function remains to be further investigated.
Overall, we identified three stress-responsive GmMYB genes: GmMYB76, GmMYB92, and GmMYB177. Overexpression of these genes in transgenic Arabidopsis plants conferred basal salt tolerance through differential regulation of a series of downstream genes. GmMYB177 and GmMYB76 may play larger roles than GmMYB92 in salt and freezing tolerance. However, the three genes appear to be negative regulators of ABA signaling. Further analysis should be performed to study the mechanisms by which the three GmMYB genes regulate plant tolerance response to various abiotic stresses.
Materials and Methods
Phylogenetic analysis of soybean MYB proteins
Previously we have identified different ESTs encoding soybean MYB transcription factors using BLASTX Network Service 39. These ESTs were used as queries to search against the data in GenBank for further confirmation. Alignments, tree construction of predicted amino acid sequences by the neighbor-joining method, and its bootstrapping (1 000 samples) were performed with CLUSTAL X (1.8). In parsimony method, the PHYLIP 3.65 package was used. Multiple most parsimonious trees were found and the consensus tree was built with the CONSENSUS program of PHYLIP.
Growth of soybean seedlings and stress treatments
Soybean (Glycine max L. Merr.) plants were grown on vermiculite under 16/8 h light/dark at 25 °C for two weeks and used for stress treatments. Soybean seedlings were carefully pulled out from vermiculite, rinsed in water, and then dehydrated on Whatman No. 3 MM paper at 25 °C with 70% humidity for various times. For ABA and salt stress treatments, two-week-old seedlings were placed hydroponically in 100 μM ABA or 150 mM NaCl for various times. Cold treatment was performed by exposing plants at 4 °C. After these treatments, the leaves of plants were collected, frozen in liquid nitrogen, and stored at −80 °C for RNA analysis.
RNA analysis, reverse transcription, and PCR (RT-PCR)
RNA isolation from soybean leaves and homozygous Arabidopsis transgenic seedlings, and RNA gel blot analysis were performed as described 78. RT-PCR was performed in 25 μl reaction volume containing 1-3 μl cDNA, 2.5 μl 10× PCR reaction buffer, 0.5 μl dNTP (10 mM/each), 0.25 μl Primer R (25 μM), and 0.25 μl Primer L (25 μM), 1 U Taq, with 30 cycles of denaturation at 94 °C for 30 s, annealing at 54-58 °C for 60 s, and extension at 72 °C for 1 min. GmMYB genes, their accession numbers, and primers used for RT-PCR analysis are listed in Supplementary information, Tables S1 and S2.
Transactivation and dimerization analysis in yeast
GmMYB76, GmMYB92, and GmMYB177 coding sequences were cloned into the DNA-BD vector pBD-GAL4 (HybriZAP-2.1 XR library construction kit and hybriZAP-2.1 XR cDNA synthesis kit; Stratagene, La Jolla, CA) to construct GAL4 DNA-BD-GmMYB fusion plasmids pBD-GmMYBs. These plasmids were transformed into the yeast strain YRG-2 harboring the LacZ and HIS3 reporter genes. The transformed yeast culture was dropped onto SD plates with or without histidine. The plates were incubated for 3 days and applied to a β-gal assay for examination of the transactivation ability.
GmMYB coding sequence was also cloned into the AD vector pAD-GAL4–2.1 to generate a GAL4-activation-GmMYB fusion plasmid pAD-GmMYB. pAD-GmMYB and pBD-GmMYB were co-transformed into the yeast strain YRG-2. The transformed yeast culture was dropped onto SD/−Trp−Leu or SD/−Trp−Leu−His plus 3-AT. The plates were incubated for 3 days and also applied to β-gal assay for examination of dimerization ability. For pBD vector construction, 5′-CCG gaa ttc ATG GAG ACC ACG AAT GTT CA-3′ and 5′-TCC ccc ggg GTG TAA ATT GCA AAA CTG GA-3′ were used for GmMYB76; 5′-ACG Cgt cga cAA ATG GGA AGG GCT CCT TGT-3′ plus 5′-AAA ctg cag TCT TTG TTG ATA TTC TTT TA-3′ or plus 5′-AAA ctg cag ATA CAT TGC CTT CAC TCT AA-3′ were used for GmMYB92 or GmMYB-N, respectively. For pAD vector construction, 5′-CGC gga tcc ATG GAG ACC ACG AAT GTT CA-3′ and 5′-CCG gaa ttc GTG TAA ATT GCA AAA CTG GA-3′ were used for GmMYB76; 5′-CGC gga tcc ATG GGA AGG GCT CCT TGT-3′ and 5′-AAA ctg cag TCT TTG TTG ATA TTC TTT TA-3′ were used for GmMYB92. For pAD and pBD vector construction, 5′-CCG gaa ttc ATG GCG ATA CAA GGC CAA AA-3′ and 5′-AAA ctg cag GAA AAG CCA ATA AGA GGG TA-3′ were used for GmMYB177.
DNA-binding ability of GmMYBs in yeast one-hybrid assay system
Three copies of each of the cis-acting DNA elements MRE1 (CCG GCA GTT AGG AT), MBSI (TAT AAC GGT TTT TTA), MRE3 (TCT AAC CTA CCA), MRE4 (TCT CAC CTA CC), and their corresponding mutant sequences mMRE1 (CCG GAA AAA AGG AT), mMBSI (TAT AAA AAT TTT TTT A), mMRE3 (TCT AAA AAA CCA), and mMRE4 (TCT CGA TCA CC) were synthesized (Sengon of Shanghai, China), annealed, and cloned into the SacI and MluI sites of reporter plasmid pHIS2, a reporter vector that contains the nutritional reporter gene HIS3. The sequence of the inserts was confirmed by sequencing. pAD-GmMYB76, pAD-GmMYB92, and pAD-GmMYB177, together with the reporter plasmid pHIS2 containing different cis-DNA elements, were co-transformed into yeast cells (Y187) and selected on selection medium. For negative Control, pAD-GmMYBs construct and pHIS2 were co-transformed into yeast cells (Y187) and growth of the transformants was examined on selection medium.
Generation of the GmMYB-transgenic Arabidopsis plants and stress treatments
The coding region of GmMYBs was cloned into the BamHI and KpnI sites of the plant expression vector pBin438 and confirmed by sequencing. The transgene was driven by the 35S CaMV promoter and an Ω translation enhancer. Transformation of Arabidopsis was performed by the vacuum infiltration method 79, using Agrobacterium tumefaciens strain GV3101. For GmMYB76, primers 5′-CGC GGA TCC GGG CAA ATT AGT ATA ATA TG-3′ and 5′-CGG GGT ACC TGC AAA GCC TAG TGT AAA TT-3′ were used. For GmMYB92, 5′-CGG GGA TCC TCT AGA GAT TAA ATG GGA AG-3′ and 5′-TAT GGT ACC GAC GAT TGC AAT GTG ATC TTC A-3′ were used. For GmMYB177, 5′-CGC GGA TCC GCC AAC CAC AGA GGA GAA GA-3′ and 5′-CGG GGT ACC CAA GGT CAT CCA GCA ACA GA-3′ were used.
For NaCl or ABA treatments, seeds (>100) of the transgenic Arabidopsis plants were planted in triplicate on filter papers saturated with different concentrations of ABA or NaCl, and incubated at 4 °C for 4 days before being placed at 22 °C for germination under a photoperiod of 16 h/8 h (light/dark). Germination (emergence of radicals) was scored after 3 days. The experiment was repeated twice and the results were consistent. Results from one set of experiments are shown.
For NaCl treatment in soil, 14-day-old seedlings of WT and transgenic seedlings from MS agar media were grown in potted soil for 7 days. Then the plants were watered with 100, 150, and 200 mM NaCl, and each concentration lasted for 4 days. The inflorescence height was measured after the treatment. The 14-day-old seedlings were also grown in pots for 7 days and then treated with high salt by placing the pots in a tray containing 600 mM NaCl. After 12 days, the survival of the plants was examined. The experiments were repeated independently three times and the results were consistent. Results from one set of experiments are shown.
For freezing tolerance assay, 14-day-old WT and transgenic seedlings from MS agar media were grown in potted soil for 7 days, and the plants were subjected to −6 °C for 2 days. After that, the plants were placed under normal condition. The survival of the seedlings was scored after 7 days and photograph was taken. The experiments were repeated independently for three times and the results were consistent. Results from one set of experiments are shown.
Determination of proline
Free proline content in plants was estimated following the methods of 80. Plant samples (100 mg) were homogenized in 1 ml of sulfosalicylic acid (3%) using a mortar, and the mixture was mixed and centrifuged at 13 000 rpm for 15 min at 4 °C. The supernatant (200 μl) was transferred to a new tube, and 200 μl of acid ninhydrin and 200 μl of acetic acid were added. The reaction mixture was boiled in a water bath at 100 °C for 1 h, and then stored at 4 °C for 30 min. After 800 μl toluene was added to extracts, the mixture was vortexed for 15 s and then 700 μl of the toluene phase was removed for measurement of the absorbance at 520 nm on a spectrophotometer. The results from three samples are averaged. Vertical bars represent standard deviation for n=3 (*P<0.05 and **P<0.01).
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
References
Klempnauer KH, Gonda TJ, Bishop JM . Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 1982; 31:453–463.
Rosinski JA, Atchley WR . Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 1998; 46:74–83.
Jin H, Martin C . Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 1999; 41:577–585.
Stracke R, Werber M, Weisshaar B . The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 2001; 4:447–456.
Romero I, Fuertes A, Benito MJ, Malpical JM, Leyva A, Paz-Ares J . More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 1998; 14:273–284.
Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 2000; 290:2105–2110.
Karpinska B, Karlsson M, Srivastava M, et al. MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol Biol 2004; 56:255–270.
Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD . A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991; 67:483–493.
Lee MM, Schiefelbein J . WEREWOLF, a MYBrelated protein in Arabidopsis, is a position dependent regulator of epidermal cell patterning. Cell 1999; 99:473–483.
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K . An Arabidopsis myb homologue is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 1993; 5:1529–1539.
Magaraggia F, Solinas G, Valle G, Giovinazzo G, Coraggio I . Maturation and translation mechanisms involved in the expression of a myb gene of rice. Plant Mol Biol 1997; 35:1003–1008.
Hoeren FU, Dolferus R, Wu YR, Peacock WJ, Dennis ES . Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 1998; 149:479–490.
Gubler F, Kalla R, Roberts JK, Jacobsen JV . Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 1995; 7:1879–1891.
Iturriaga G, Leyns L, Villegas A, Gharaibeh R, Salamini F, Bartels D . A family of novel myb-related genes from the resurrection plant Craterostigma plantagineum are specifically expressed in callus and roots in response to ABA or desiccation. Plant Mol Biol 1996; 32:707–716.
Yang Y, Klessig DF . Isolation and characterization of a tobacco mosaic virus-inducible myb homologue from tobacco. Proc Natl Acad Sci USA 1996; 93:14972–14977.
Tamagnone L, Merida A, Parr A, et al. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate Phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998; 10:135–154.
Newman LJ, Campbell MM . MYB proteins and xylem differentiation. In: Savidge R, Barnett J, Napier R, eds. Cell and molecular biology of wood formation. Oxford: BIOS Scientific Publishers, 2000:437–444.
Lauvergeat V, Rech P, Jauneau A, Guez C, Coutos TP, Grima-Pettenati J . The vascular expression pattern directed by the Eucalyptus gunnii cinnamyl alcohol dehydrogenase EgCAD2 promoter is conserved among woody and herbaceous plant species. Plant Mol Biol 2002; 50:497–509.
Oh S, Park S, Han KH . Transcriptional regulation of secondary growth in Arabidopsis thaliana. J Exp Bot 2003; 54:2709–2722.
Patzlaff A, McInnis S, Courtenary A, et al. Characterisation of a pine MYB that regulates lignification. Plant J 2003; 36:743–754.
Newman LJ, Perazza DE, Juda L, Campbell MM . Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J 2004; 37:239–250.
Cheong YH, Moon BC, Kim JK, et al. BWMK1, a rice itogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol 2003; 132:1961–1972.
Shinozaki K, Yamaguchi-Shinozaki K . Gene expression and signal transduction in water-stress response. Plant Physiol 1997; 115:327–334.
Chen TH, Murata N . Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol 2002; 5:250–257.
Zhang X, Fowler SG, Cheng H, et al. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J 2004; 39:905–919.
Nakashima K, Yamaguchi-Shinozaki K . Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol Plant 2006; 126:62–71.
Thomashow MF . Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Plant Mol Biol 1999; 50:571–599.
Zhu JK . Salt and drought stress signal transduction in plants. Ann Rev Plant Biol 2002; 53:247–273.
Shinozaki K, Yamaguchi-Shinozaki K, Seki M . Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 2003; 6:410–417.
Yamaguchi-Shinozaki K, Shinozaki K . Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 2005; 10:88–94.
Narusaka Y, Nakashima K, Shinwari ZK, et al. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 2003; 34:137–148.
Kim JC, Lee SH, Cheong YH, et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J 2001; 25:247–259.
He CY, Zhang JS, Chen SY . A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor Appl Genet 2002; 104:1125–1131.
Umezawa T, Mizuno K, Fujimura T . Discrimination of genes expressed in response to the ionic or osmotic effect of salt stress in soybean with cDNA-amplified fragment length polymorphism. Plant Cell Environ 2002; 25:1617–1625.
Liao H, Wong FL, Phang TH, et al. GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 2003; 318:103–111.
Aoki A, Kanegami A, Mihara M, Kojima T, Shiraiwa M, Takahara H . Molecular cloning and characterization of a novel soybean gene encoding a leucine-zipper-like protein induced to salt stress. Gene 2005; 356:135–145.
Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, Chen SY . Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet 2005; 110:1355–1362.
Luo GZ, Wang HW, Huang J, et al. A putative plasma membrane cation/proton antiporter from soybean confers salt tolerance in Arabidopsis. Plant Mol Biol 2005; 59:809–820.
Tian AG, Wang J, Cui P, et al. Characterization of soybean genomic features by analysis of its expressed sequence tags. Theor Appl Genet 2004; 108:903–913.
Kranz HD, Denekamp M, Greco R, et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 1998; 16:263–276.
Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK . Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J Biol Chem 2000; 275:24208–24214.
Martin C, Paz-ares J . MYB transcription factors in plants. Trends Genet 1997; 13:67–73.
Seong HA, Kim KT, Ha H . Enhancement of B-MYB transcriptional activity by ZPR9, a novel zinc finger protein. J Biol Chem 2003; 278:9655–9662.
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF . Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 2004; 40:22–34.
Uimari A, Strommer J . MYB26: a MYB-like protein of pea flowers with affinity for promoters of Phenylpropanoid genes. Plant J 1997; 12:1273–1284.
Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B . Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol 1998; 36:741–754.
Sugimoto K, Takeda S, Hirochika H . MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 2000; 12:2511–2528.
Wanner LA, Junttila O . Cold-induced freezing tolerance in Arabidopsis. Plant Physiol 1999; 120:391–399.
Vannini C, Locatelli F, Bracale M, et al. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 2004; 37:115–127.
Yamaguchi-Shinozaki K, Shinozaki K . A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 1994; 6:251–264.
Liu Q, Sakuma H, Abe M, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998; 10:1391–1406.
Shinozaki K, Yamaguchi-Shinozaki K . Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 2000; 3:217–223.
Yoshiba Y, Kiyosue T, Katagiri T, et al. Correlation between the induction of a gene for Al-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 1995; 7:751–760.
Strizhov N, Abraham E, Okresz L, Blickling S, Zilberstein A . Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J 1997; 12:557–569.
Gilmour SJ, Artus NN, Tomashow MF . cDNA sequence analysis and expression of two cold-regulated genes Arabidopsis thaliana. Plant Mo1 Biol 1992; 18:13–21.
Welin BV, Olson A, Palva ET . Structure and organization of 2 closely-related low-temperature-induced Dhn/Lea/Rab-like genes in Arabidopsis thaliana L Heynh. Plant Mol Biol 1995; 29:391–395.
Horvath DP, McLarney BK, Thomashow MF . Regulation of Arabidopsis thaliana L.(Heyn) cor78 in response to low temperature. Plant Physiol 1993; 103:1047–1053.
Nordin K, Vahala T, Palva ET . Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 1993; 21:641–653.
Lin C, Thomashow MF . DNA sequence analysis of a complementary DNA for cold-regulated Arabidopsis gene cor15 and characterization of the COR15 polypeptide. Plant Physiol 1992; 99:519–525.
Koornneef M, Reuling G, Karssen C . The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Plant Physiol 1984; 61:377–383.
Kurkela S, Borg-Franck M . Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol 1992; 19:689–692.
Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K . Characterization of cDNA for a dehydration-inducible gene that encodes a CLP A, B-like protein. Biochem Biophys Res Commun 1993; 196:1214–1220.
Lang V, Palva ET . The expression of a Rab-related gene, Rab18, is induced by abscisic-acid during the cold-acclimation process of Arabidopsis thaliana (L) Heynh. Plant Mol Biol 1992; 20:951–962.
Weston K . Myb proteins in life, death and differentiation. Curr Opin Genet Dev 1998; 8:76–81.
Jin H, Cominelli E, Bailey P, et al. Transcriptional repression by At-MYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 2000; 19:6150–6161.
Hemm MR, Herrmann KM, Chapple C . AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci 2001; 6:135–136.
Villalobos MA, Bartels D, Iturriaga G . Stress tolerance and glucose insensitive phenotyes in Arabidopsis overexpressing the CpMYB10 transcription factor gene. Plant Physiol 2004; 135:309–324.
Zhu JH, Verslues PE, Zheng XW, et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 2005; 102:9966–9971.
Nagaoka S, Takano T . Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot 2003; 54:2231–2237.
Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM . AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol 2005; 15:1201–1206.
Cominelli E, Galbiati M, Vavasseur A, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 2005; 15:1196–1200.
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K . Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003; 15:63–78.
Sakuma Y, Maruyama K, Osakabe Y, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006; 18:1292–1309.
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K . Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 1999; 19:287–291.
Delauney AJ, Verma DPS . Proline biosynthesis and osmoregulation in plants. Plant J 1993; 4:215–223.
Xin ZG, Browse J . Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA 1998; 95:7799–7804.
Khedr AHA, Abbas MA, Wahid AAA, Quick WP, Abogadallah GM . Proline induces the expression of salt-stress-responsive proteins and may improve the adaptation of Pancratium maritimum L. to salt-stress. J Exp Bot 2003; 54:2553–2562.
Zhang JS, Gu J, Liu FH, Chen SY . A gene encoding a truncated large subunit of Rubisco is transcribed and salt-inducible in rice. Theor Appl Genet 1995; 91:361–366.
Bechtold N, Pelletier G . Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 1998; 82:259–266.
Bates LS, Waldre RP, Teare ID . Rapid determination of free proline for water stress studies. Plant Soil 1973; 39:205–208.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (30490254, 30671316), the National Basic Research Program of China (2006CB100102), and the Hi-Tech Research and Development Program of China (2006AA10Z113, 2006AA10A111).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary information, Figure S1
Phenotype of the transgenic plants overexpressing the GmMYB76, GmMYB92 or GmMYB177 compared to Col under normal growth condition. (PDF 153 kb)
Supplementary information, Table S1
The GmMYB gene family in soybean plants. (PDF 15 kb)
Supplementary information, Table S2
Primers used for examination of GmMYB gene expression by RT-PCR (PDF 18 kb)
Rights and permissions
About this article
Cite this article
Liao, Y., Zou, HF., Wang, HW. et al. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res 18, 1047–1060 (2008). https://doi.org/10.1038/cr.2008.280
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2008.280
Keywords
This article is cited by
-
Potential Breeding Strategies for Improving Salt Tolerance in Crop Plants
Journal of Plant Growth Regulation (2023)
-
Twenty years of mining salt tolerance genes in soybean
Molecular Breeding (2023)
-
Genome-wide characterization of Alfin-like (AL) genes in apple and functional identification of MdAL4 in response to drought stress
Plant Cell Reports (2023)
-
Gene regulation network analyses of pistil development in papaya
BMC Genomics (2022)
-
Genome-wide identification and expression analysis of the bZIP transcription factor family genes in response to abiotic stress in Nicotiana tabacum L.
BMC Genomics (2022)