Abstract
We applied the technique of mRNA differential display to normal liver tissue and hepatoma cell line Hep3B. One of the isolated cDNA clones was expressed in human normal liver tissue but not in the human hepatocarcinoma cell line. Northern Blot analysis confirmed that high level of mRNA was expressed in human normal liver tissue but the level was decreased in non-cancerous liver tissue from hepatoma patients. Low level or no expression was observed in human hepatoma tissue. One of these transcripts was about 1.8 kb in length. Southern Blot analysis showed that it was a single copy gene. We obtained a full length cDNA clone of 2,395 bp by screening human liver 5′-stretch plus cDNA library. Nucleotide sequence indicated that this clone was highly homologous to aryldialkyl-phosphatase and possessed two polymorphic sites. Aryl-dialkyl-phosphatase which has a prominent role in the metabolism of several toxic, synthetic compounds, may be potentially related to human hepatocarcinoma susceptibility. The biological significance of its differential expression in normal versus malignant tissue is discussed.
Similar content being viewed by others
Introduction
Cancer is believed to be the result of cumulative multiple genetic aberrations and a process that may operate through either of two different mechanisms. The first mechanism involves a class of oncogenes and tumor suppressor genes, whose activation and/or inactivation is directly involved in tumorigenesis. The second mechanism involves a class of cancer susceptibility genes, which influence the rate at which mutations occur and/or accumulate. Cancer susceptibility genes are involved in DNA repair, phase I and II metabolism and so on. Variation of genes from class II, while not directly responsible for transformation, may act to alter genes categorized in class I. The aberrant expression of these critical genes and their downstream subsets of genes might enable a cell to proliferate out of control. To identify and characterize as many of these genes as possible has become crucial to understand the nature of the disease and to devise rational therapies and prevention.
Several important genes implicated in tumorigenesis have been isolated1, 2 by mRNA differential display (mRNA DD) developed by P. Liang and A. Pardee in19923, 4. In 1993, our laboratory, in collaboration with Dr. Takis Papas, FCRDC, NCI, applied mRNA DD to identify and isolate a small subset of genes that are differentially expressed in normal liver tissue versus human hepatoma cell line Hep3B. By this method we have obtained 28 differentially expressed clones5. In this paper we report that one of these clones that is overexpressed in normal liver tissue and underexpressed in hepatocarcinoma tissue.
Materials and Methods
Materials
Human liver 5′-stretch plus cDNA library was provided by CLONTECH Laboratories, Inc. Human primary hepatoma and corresponding non-cancerous liver tissues were collected from Qidong Cancer Institute. All the specimens were confirmed by histopathological examination. Nick translation DNA labeling kit, random primer labeling kit, Geneclean DNA purification kit and USB sequenase version 2.0 kit was obtained respectively from Boehringer Mannheim, Stratagene, BIO101 and United States Biochemical Corporation. 32p-(α)-dCTP, 32P-(α)-dATP and 35s-(α)-dATP were obtained from Amersham Corporation. Restriction enzymes, DNA ligase, DNase and RNase were obtained from Promega, Boehringer Mannheim, Sino-American Biotechnology Company etc..
Screening of cDNA library
Screening of cDNA library was performed with 32P-(α)-dCTP labeled C2L2 probe at high stringency, hybridized overnight at 42 °C in 50% formamide / 6 × standard saline sodium citrate (SSC) / 5 × Denhardt's solution / 0.5% SDS / 200 mg of salmon sperm DNA and washed in 0.5 × SSC / 0.1% SDS at 42 °C , 30 min, at 55 °C , 1 h. After the third round of screening, twelve independent clones were isolated. The phage DNA was purified by liquid amplification and digested with EcoRI (cloning site). The inserts were confirmed by Southern blot and then subcloned into the pBluescript SK(−) vector
Northern and Southern blot analysis
Total RNA was isolated from frozen samples of human hepatic cancer and non-cancerous liver tissue by extraction with guanidine isothiocyanate-phenol and ethanol precipitation. DNA was prepared from frozen human liver tissues by extraction with phenol, phenol-chloroform and purified by ethanol precipitation. Southern and Northern transfer were carried out with standard teehniques. Hybridization with insert cDNA fragment described above labeled with 32P-(α)-dCTP at high stringency was done overnight at 42 °C in 50 % (vol/vol) formamide / 6 × SSC / 5 × Denhardt's solution / 0.5% SDS / 200 μ g of salmon sperm DNA and washed in 0.5 × SSC / 0.1% SDS at 42 °C , 30 min, at 65 °C , 1 h, followed by autoradiography at −70 °C .
Nucleotide sequencing and sequence analysis
Dideoxynucleotide chain termination method was employed with the 35S-(α)-dATP (Amersham > 800 ci/mmol) according to the USB Sequenase Version 2.0 Kit manual. A program for analyzing DNA sequences was performed with HIBIO DNASIS™ and a program for analyzing protein sequences with HIBIO PROSIS™.
Result
Isolation of C2L2 clone
Human liver 5′-stretch plus cDNA library was screened by using C2L2 probe. After three rounds of screening, twelve positive phages that gave the strongest signal were purified by liquid amplification. Inserts were cleaved by restriction enzyme EcoRI, confirmed by subsequent Southern blot hybridization with C2L2 probe and finally subcloned into pBluescript SK(−) vector (Fig 1).
Northern blot analysis
Northern blot analysis was carried out in 4 samples of normal human liver tissue and in 6 pairs of samples from human primary hepatic cancer and its surrounding non-cancerous hepatic tissues. It was demonstrated that this gene was expressed more strongly in normal liver tissues than in surrounding non-cancerous hepatic tissues, and its expression was low or absent in hepatoma tissues. Moreover, a single hybridization band was observed, corresponding to a transcript of approximately 1.8 kb (Fig 2).
Southern blot analysis
Genomic DNA extracted from human liver, hepatic cancer and surrounding noncancerous tissues was digested with PstI and EcoRI respectively. Fig 3 showed that only one hybridization band was observed at about 3 or 4 kb respectively in each restriction digested lane. These data suggested that this gene was probably of a single copy entity. The positive bands from both normal liver and hepatoma tissues were located at the same position. These findings indicated that C2L2 gene was not lost or grossly aberrant in hepatic cancer tissue, but the existence of subtle genomic alterations could not yet be excluded from the present data.
Sequencing of the C2L2 cDNA clone
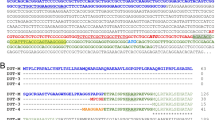
The C2L2 cDNA clone contained 2,395 base pairs, covering the entire protein coding sequence (Fig 4). Fifty-four nucleotides preceded the codon ATG which initiated an open reading frame coding for 355 amino acids. A stop codon, TAA, was followed by an additional 1,276 nucleotides of 3′ non-coding sequence. The ATG at position 1 was likely the start position since there was a stop codon beginning 36 nncleotides upstream from this ATG. Poly(A) signal (AATAAA) was located at nt 2,052. There were two polymorphic sites in the sequence. One was located at nt217 with A or T (Fig 5A), and the other at nt 629 with G of A (Fig 5B).
The full cDNA sequence of C2L2. The coding protein had 355 amino acids. The poly(A) tailing sequence was located in nt 2,052 bp. Two polymorphic sites were located in nt 217 bp (A or T), nt 629 bp (G or A) respectively. The nucleotide sequence appeared in the GenBank, EMBL and DDBJ nucleotide sequence databases with the following accession number U53784, Z70723, D84371.
Analysis of DNA sequence
Comparing C2L2 cDNA sequence with the databases in the GenBank and EMBL, we found the major sequence was highly homologous to aryl-dialkyl-phosphatase(Fig 6). Aryl-dialkyl-phosphatase was a gene with two polymorphic sites6, 7 in accord with our results. There were three kinds of reported sequence8, whose length was 1576, 1217, 1337 bp respectively. They were all highly homologous to C2L2, but the sequence of C2L2 was the longest, which was 2,395 bp. Because C2L2 and these matched cDNA clones shared the same coding sequence, we concluded that they were the same gene.
Protein database searches of C2L2 indicated that N-terminal sequences showed similarity to other protein secretion signal peptide sequences (Fig 7). Conservation of specific amino acids was apparent, but particularly interesting were the conserved three amino acid residues LAL that were contained by some protein sequences in their hydrophobic core region. Hydrophobicity analyses performed with PROSIS software according to the Hopp algorithm clearly showed the hydrophobic amino termini of C2L2 (Fig 8).
Discussion
mRNA DD technique applied to isolate differentially expressed genes
To identify and isolate those genes that are differentially expressed in normal tissue versus tumor tissue is crucial to an understanding of the genetic aspect of tumorigenesis. The mRNA DD technique offers a novel, rapid method to this research. In spite of many advantages of mRNA DD in the analysis of altered gene expression, the major disadvantage of this technique is to generate high numbers of 'false positives'. In our experiment, we find two out of twenty-eight clones are 'true positive' and one of them is to be further characterized. These artifacts could be caused due to the following reasons: firstly, PCR itself may give some artifact results. Secondly, cDNA recovered from what may appear to be a unique band may contain additional, undeteetable overlapping, or unresolved cDNA. Contaminating cDNA might have been copurified with differentially displayed cDNA. Thirdly, some observed bands arise as a result of the amplification of cDNA sequences primed by only one of the two primers used in the PCR. This may result from the A-T rich nature of the 3′ untranslated region of many genes, which can be recognized by the anchored primer. Besides, the sequence of arbitrary and its inverse-complement may exist in the same cDNA, resulting in the amplification of a region of cDNA between these two positions. Now an improved mRNA DD technique9 is being established for analyzing differentially expressed cDNA clones from normal liver and hepatic cancer tissue.
The genetic polymorphism of the poison metabolizing enzymes—one of the important aspects of genetic susceptibility to cancer
Only a small number of individuals exposed to any given level of carcinogens will develop cancer. Interindividual differences in the susceptibility to these environmental exposures have been postulated. In fact, almost each phase of the multistage processes of carcinogenesis may be putatively modified by inherited or acquired interindividual differences in susceptibility. There are many demonstrable interindividual differences in the metabolic capability to convert proearcinogens to carcinogens and to detoxify carcinogens. These genetically determined differences result in variations in host susceptibility10. Polymorphic genes that encode for human biotransformation enzymes and affect the rate of metabolism of certain drugs and xenobiotics have been identified as one determinant of the susceptibility to environmental toxic chemicals. For example, GST M1 is an important phase II biotransformation enzyme and it detoxifies AFB1-2, 3-epoxide, which is the terminal carcinogen of AFB1. GST M1 possesses three genotypes: GST M1a, GST M1b, GST M1-null11. Individuals with a homozygous GST M1-null type express no protein so that they do not have the ability to detoxify AFB1-2, 3-epoxide. It has been suggested that this genotype should be associated with an increased risk of hepatic cancer. Recently Sheng et al have reported that N-acetyltransferase involved in detoxifying a number of poisons is associated with the susceptibility of hepatic cancer12. But there is no conclusion yet about which carcinogen is involved in hepatocarcinogenesis.
Aryl-dialkyl-phosphatase is a kind of esterase with a broad substrate specificity13 Its substrates include aromatic carboxylic acid esters, carbamic esters and esters of phosphorus acid. This kind of esterase may have a prominent role in the metabolism of several toxic, synthetic compounds. But its endogenous substrates remain unknown and its physiological role has not yet been identified. According to results from our laboratory and other laboratories, aryl-dialkyl-phosphatase is also a type of polymorphic poison metabolic enzyme6, 7. Three phenotypes (Type A, Type B, Type AB) have been found to be controlled by two major alleles at a single autosomal locus. Sequencing of the coding region revealed that an Arg/Arg homozygote is associated with the high-activity phenotype (Type A), a Gln/Gln with the lowactivity phenotype (Type B), and an Arg/Gln heterozygote with the intermediate phenotype (Type AB). When the phenotype of this esterase is of low-activity, the ability to detoxify poison decreases significantly. According to long-term studies, the well-established environmental triggers for hepatocarcinoma include hepatitis B virus (HBV), hepatitis C virus (HCV), aflatoxin, nitrosamine, microcystin, ethanol and so on. At present, we are not sure whether other environmental chemical carcinogens might be involved in hepatocarcinogenesis and their metabolic processes do require aryl-dialkyl-phosphatase. However, a recent report has described that the development of hepatocarcinoma may be linked to the excessive exposure of organophosphate. As we described above, the aryl-dialkyl-phosphatase may be suppressed in its expression in hepatic cancer tissue as well as the non-cancerous liver tissue. It could be postulated that the downregulated expression of this enzyme in liver cells due to some factors (viral or others) could lead to the impairment of metabolic defensive mechanism to detoxify the organophosphate toxins, thereby rendering liver cells susceptible to carcinogen challenge. Moreover, an alternative hypothesis of the impairment in the esterase-dependent immune surveillance system related to the development of lymphoid tumor was recently postulated. It is suggested that the development of lymphoma is correlated to the exposure of organophosphorus compounds. This hypothesis puts emphasis on the possible new mechanism of organophosphorus compound that may impair the esterase-dependent host immune system as well as cause chromosome aberration. Thereby the host cells are more susceptible to viral carcinogenesis. The latter postulation may give some insight in search for the correlation between regulation of aryl-dialkyl-phosphatase to detoxify organophosphorus compounds and the impairment of host immune system in human hepatocarcinogenesis.
References
Sager R, Anisowicz A, Neveu M, Liang P, Sotiropoulou G . Identification by differential display of alpha 6 integrin as a candidate tumor suppressor gene. The FASEB Journal 1993; 7:964–70.
Zhang LX, Medina D . Gene expression screening for specific genes associated with mouse mammary tumor development. Molecular Carcinogenesis 1993; 8:123–6.
Liang P, Averboukh L, Keyomarsi K, Sager R, Pardee AB . Differential display and cloning of messenger RNAs from human breast cancer versus mammary epithelial cells. Cancer Res 1992; 52:6966–8.
Liang P, Pardee AB . Differential display of eukaryotic messenger RNA by means of the polymerase chain reation. Science 1992; 257:967–71.
Da Fang Wan, Xue Li Zhang, Xiao Mei Zhou, Jian Ren Gu . cDNA clones of differential expression between human normal liver tissue and hepatocarcinoma. National Medical Journal of CHINA 1995; 75(10):606.
Eckerson HW, Wyte CM, La Du BN . The human serum paraoxonase/arylesterase polymorphism. Am J Hum Genet 1983; 35:1126–38.
Adkins Steve, Gan Karen N, Mody Malay, et al. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am J Hum Genet 1993; 52:598–608.
Hassett C, Richter RJ, Humber R, et al. Charaterization of cDNA clones encoding rabbit and human serum paraoxonase: the mature protein retains its signal sequence. Biochemistry 1991; 30:10141–9.
Liang P, Zhu W, Zhang XY, Guo Z, P.O'Connell R, Averboukh L, Wang FL and Pardee AB . Differential display using one-base anchored oligo-dT primers. Nucleic Acids Res 1994; 22(25) 5763–4.
Spitz MR, et al. Genetic susceptibility to cancer. Cancer 1993; 72(3):991–5.
Laisney V, Cong NV, Gross MS, et al. Human genes for glutathione S-transferases. Human Genet 1984:68–221:
Zhao You Tang, Sheng Long Ye . Recent progress in liver cancer and hepatitis International Academic Publishers 1996; Page 66–67.
Mackness MI . Commentary: A-esterase: enzymes looking for a role. Biochem Pharmacol 1989; 38:385–90.
S. Cordier, Le Thi Bich Thuy, P. Verger, et al. Viral infections and chemical exposures as risk factors for hepatocellular carcinoma in Vietnam. Int J Cancer 1993; 55:196–201.
David S . Newcombe Immune surveillance, organphosphorus exposure, and lymphomagenesis Lancet 1992:339:539–41.
Acknowledgements
This work was supported by grants from the Foundation of National Pandeng Project and the Foundation of Human Genome Project. The nucleotide sequence data reported in this paper appeared in the Gen Bank, EMBL, and DDBJ nucleotide sequence databases with the following accession number U53784, Z70723, D84371.
Author information
Authors and Affiliations
Corresponding author
Additional information
*This paper is specifically dedicated to Professor Lu Ji SHI on the occasion of his 80th birthday
Rights and permissions
About this article
Cite this article
Wang, K., Wan, D., Qiu, X. et al. Differential expression of a cDNA clone in human liver versus hepatic cancer highly homologous to aryl-dialkylphosphatase. Cell Res 7, 79–90 (1997). https://doi.org/10.1038/cr.1997.9
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1997.9