Abstract
The somaclone, C39, derived by tissue culture from the obligate apomict Paspalum dilatatum cv Raki (2n=50), had 50 chromosomes and a karyotype apparently identical to Raki. SC2 seedlings of C39 showed a high degree of phenotypic variation which was often associated with increased chromosome numbers, but some of the variant seedlings were karyotypically indistinguishable from Raki or C39. Plants with increased chromosome numbers exhibited a high degree of intraplant chromosome variation (aneusomaty). In one of the SC2 seedlings, the chromosome number of root tip cells varied from 58 to 82 and in several other seedlings the range was more than 10. The results suggested that the ability to form seed apomictically was much reduced in C39 and that this plant showed some capacity for sexual reproduction and the resulting seedlings, with a chromosome number of about 70, were genetically unstable. Of 11 SC2 seedlings examined cytologically, 6 did not produce any viable seed. Seedlings grown from seed of the remaining 5 plants showed that aneusomaty persisted in the SC3 generation. SC3 seedlings which were phenotypically similar to their maternal parent showed a similar range of chromosome numbers to that parent. Some of the SC3 seedlings exhibited an even wider range of chromosome numbers (e.g.56-136), and these plants were all dwarfs.
Similar content being viewed by others
Introduction
Paspalum dilatatum Poir. is an important forage plant in many subtropical and warm temperate countries. It is a complex hybrid with chromosomes contributed from three species, the probable parent species being P. intermedium and P. jurgensii both contributing 20 chromosomes, and an as-yet-unidentified species contributing 10 chromosomes1. Meiosis is irregular as there are 20 bivalent and 10 univalent chromosomes2. Apomixis allows the plant to produce seed with a stable chromosome complement of 50 but seed germinability based on total florets produced is less than 25%3. Though likely to have been important in the evolution of the species, apomixis is a barrier to improvement by conventional breeding4. Hybridization between sexual and apomictic plants has been an effective way to break apomixis in some grasses4. However, the only sexual biotype of P. dilatatum which has been identified is a tetraploid (2n=40) and this biotype has very low crossability with the common apomictic biotype5, 6. Interspecific crosses and genetic modification by exposure to radiation have also failed to produce useful genetic variation7, 8, 9, 1.
It has been shown that regeneration of plants from callus cultures can induce variations which may be useful for breeding purposes10, 11. In Paspalum dilatatum cv Raki Davies and Cohen(1992) regenerated a large number of plants (somaclones) from embryogenic callus induced on the scutellum of immature embryos. Extensive phenotypic variation in vigor, growth habit, leaf width, and leaf chlorophyll concentration were exhibited by the somaclones obtained (SC1 generation) and most had greatly reduced seed viability compared with Raki. Some of the somaclones produced seedlings(SC2 generation) that were also highly variable and this variation reappeared to a lesser extent in the next seed generation (SC3). This variation was unexpected as it was anticipated that the somaclones would produce uniform SC2 offspring by apomixis because they were derived from an apomictic donor plant. Davies and Cohen(1992) suggested that the unexpected variation in SC2 seedlings could have been due to a breakdown in the apomixic pathway to seed production in the somaclones, and that SC2 seed may have arisen by a sexual pathway. If sexual reproduction in the somaclones can be proved it follows that tissue culture is a means of breaking apomixis in P. dilatatum.
Meiosis is irregular in P.dilatatum so that if somaclones reproduced sexually the resulting SC2 seedlings could be expected to vary in chromosome number. The detection of changes in chromosome number in the SC2 seedlings would therefore be a useful first step in determining whether the somaclones reproduced sexually. In this paper we present the results of a cytogenetic study of P. dilatatum cv Raki, one of the somaclones, C39, and some of the progeny from two subsequent seed generations developed from C39. The somaclone C39 was chosen because it produced viable seed, albeit very much reduced compared with Raki, and its progeny varied in phenotype with some approaching Raki in vigor and seed viability.
Materials and Methods
Seedlings of Raki and the primary somaclones (SC1) were grown in the field for growth studies for two seasons12. Seedlings of the SC2 generation were germinated in a greenhouse and transplanted into the field for observation over two seasons. For the cytological studies, tillers with roots were transplanted into 15cm pots which were grown in a greenhouse.
Root tips were collected from the following plants.
(a) seedlings of Paspalum dilatatum cv Raki.
(b) C39, a somaclone derived from Raki.
(c) 11 SC2 seedlings grown from open pollinated seed produced by C39. These plants were representative of the variation found in this generation.
(d) 16 SC3 seedlings, consisting of 3 or 4 seedlings from each of the 5 SC2 seedlings which produced viable seed(Tab 1). The SC3 seedlings were from open pollinated seed. In the SC3 generation, 76-92% of the seedlings were similar to their maternal parent, the remainder being off-types12. In 4 of the 5 SC3 families, (seedlings of C39 /1, /4, /5 and/10) two maternal seedlings and one off-type seedling were selected for study. For the fifth SC 3 family (seedlings of the dwarf C39 /2), one maternal seedling, one seedling similar to the SC2 parent except with pale-green leaves, and 2 very low vigor seedlings were selected. Phenotypic details of these plants axe presented in Tab 1 where plants with maternal characteristics are indicated by an asterisk.
Roots were collected from plants during the period from September 1985 to December 1988. Root tips were pretreated in 0.05 % colchicine for 4-6 h then fixed overnight in a 3:1 mixture of absolute ethanol and glacial acetic acid at 4 °C. The roots were then treated with 2% macerase, 2% cellulase (pH 4.8) at 37 °C for 1-1.5 h. The softened root tips were macerated and stained in modified carbol fuchsin13 and then squashed under coverslips which were sealed with nail polish to make temporary slides. Later the cover slips were removed by freezing the slides in liquid air or dry ice. The slides were then dehydrated in absolute ethanol and mounted in DPX.
From these slides a number of well spread mitoses were photographed and the chromosomes were counted. Between 16 and 72 counts were made for each plant; this involved counting squashes prepared from 3 to 12 roots. In total, counts were made on 1238 mitoses from 144 roots. The numbers of samples examined from each plant are listed in Tab 2.
From the photographs 83 kaxyotypes were analysed in detail. From three to fifteen karyotypes per plant were made from Raki, C39, the SC2 plants and one SC3 plant. The karyotypes were prepared from well spread mitoses in mid prophase to early metaphase. Chromosome length and centromere position as well as chromosome shape and staining pattern (natural banding) were used to arrange the chromosomes.
Results
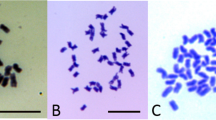
The same number of chromosomes (2n=50) were counted in 57 mitoses from Raki and in 26 mitoses from C39. Furthermore, the karyotype of these plants appears to be identical (Fig 1a, 1b) even though the two plants were phenotypically different (Tab 1).
Karyotypes from four plants. (a) The original parent, Raki, showing 20 pairs of larger chromosomes plus 10 smaller chromosomes. (b) Somaclone C39, regenerated by callus tissue culture from Raki, with a chromosome number identical to Raki (2n=50). (c) SC2 seedling, C39/11, with a reduced chromosme number (2n=48) and showing a translocation from a small chromosome to the short arm of a long chromosome (arrow). (d) SC2 seedling, C39/3, with an increased number of chromosomes. This example has 76 chromosomes, with examples of trisomy(*) and tetrasomy (•) and 17 additional small chromosomes (underlined). Scale bar 10 μm.
The chromosome numbers of the 11 SC2 seedlings fell into three groups. In one group, four plants (C39 /4, / 7, / 8, / 9) had the same chromosome number (2n=50) and their karyotypes were indistinguishable from Raki and C39. C39 / 4 had a similar phenotype to C39, except for improved seed set. The other 3 SC2 seedlings with 50 chromosomes, (C39 /7, / 8 and / 9) varied in phenotype and none of these seedlings produced viable seed.
A second group of six seedlings (C39/ 1, / 2, /3, / 5, /6, /10) showed an increase in chromosome number of about 20 compared with Raki and C39. Another striking feature was the considerable variation in chromosome number between roots and even between cells of the same root. This variation was not a short-term instability in which chromosomes were gradually lost. The range of chromosome numbers recorded in root tips collected from the same plant in two successive years was identical (Fig 2). The variation in chromosome number was not a simple case of chimerism between cells of different chromosome number as the variation in an individual plant ranged from 9 (74-82) in C39 /3, to 25 (58-82) in C39/ 6 (Tab 2) . In this group of six seedlings there were three different phenotypes. C39/1, /5 and / 10 were three high vigor plants with the same phenotype (wider leaves, coarser stems and higher seed viability than C39) and similar modal ranges of about 66-76 chromosomes (Tab 2). C39/6 had very low vigor, yellow anthers and did not produce any viable seed. The modal range of this plant was 72-74 chromosomes. C39/2 was also of low vigor but had purple anthers and higher seed viability than C39. This plant had a modal range of 74-78 chromosomes. Although C39/3 was very similar to C39/2 in vigor and leaf width and had similar modal chromosome numbers, it produced no viable seed.
An unusual prostrate plant, C39 /11, was in a group of its own with a modal chromosome number of 48, the only SC2 plant studied with less than 50 chromosomes.
Karyotypes prepared for the 7 SC2 plants in which variation was detected were analysed in detail. Of the 34 mitoses examined from C39 /11, 32 were less than diploid (2n=<50). A karyotype showing this is shown as Fig 1c. In the other SC2 plants, apparent instances of trisomy, tetrasomy, pentasomy and one of octosomy were observed. Additional small chromosomes of various sizes were also observed in many cells with the number ranging from 12-20. Examples of such variations are apparent in the karyotype derived from one root tip of C39/3 and shown as Fig 1d. The additional small chromosomes were not greatly reduced in size and so were unlikely to be B chromosomes. The frequency of polysomy among the 36 karyotypes of SC2 plants examined appeared to be very high for two distinctive pairs of submetacentric chromosomes (underlined in Fig 1 d).
Chromosomal variations were also observed in SC3 progeny from the five SC2 plants which produced viable seeds (C39 /1, 2, 4, 5, 10). SC3 plants with the same phenotype as their maternal SC2 parent also had chromosome counts similar to that parent (Tab 2, Fig 3, 4).
SC3 plants which differed in phenotype from their SC2 parent also differed in chromosome number. Three SC2 parents (C39 /1,5,10) produced a few low vigor /narrow leafed plants, distinctly different from the maternal parent. The distribution of chromosome numbers in C39 /1 /4 and C39 /5/3 were bimodal with one group of cells containing more than 90 chromosomes and a second group of cells containing less than 80 chromosomes (Tab 2, Fig 3). In C39/10 /8 most of the cells had more than 90 chromosomes with the remaining cells having 56-86 chromosomes (Tab 2, Fig 4).
The pale-green leafed seedling C39 /2/ 2 had a modal chromosome number of 54 (Fig 5) compared with a modal range of 74-78 chromosomes in both C39 /2 and C39 /2/1 (Tab 2). This was the largest drop in chromosome number between an SC3 plant and its maternal parent. Plants C39 /2 /4 and C39 /2 /5 were both extreme dwarfs of very low vigor with modal chromosome ranges of 66-70 and 70-74 respectively, but 2 cells with 134 and 136 chromosomes were found in C39 /2/5.
C39 /4 was a plant with a stable 50 chromosome complement in the SC2 generation but produced seedlings with variable chromosomes in the SC3 generation. One of these plants C39 /4 /2 had a maternal phenotype and only a small proportion of cells showed an increased chromosome number. However, C39 /4 /8 was a dwarf with a modal chromosome range of 74-78.
Discussion
There have been reports of somaclones of wheat and tomato with some variation in chromosome number, but their seed progeny had a normal chromosome complement14, 15. In Panicum maximum, an apomictic plant, no variation in either phenotype or chromosome number was found in either the primary somaclones or their seedling progeny16. In another apomictic grass, Pennisetum americanum, plants regenerated from scutellar callus were mostly stable diploids but a few aneuploid and polyploid plants were found17.
The results of the present work differ from the above reports in two significant respects. Firstly, the wide variation in chromosome number that was found within individual plants and secondly, the chromosome number variation between plants in the SC2 and SC3 generations.
Within plant variation
In the results reported above, chromosome instability was found in plants 2-4 years from seed and the range of chromosome numbers was the same over at least two successive years (Fig 2). Furthermore, plants were found to pass on this trait to progeny, presumably by apomictic development of embryo sacs from nucellar tissue. Since embryo sacs arise from a single cell, the variation in chromosome number would appear to arise de novo in somatic cells.
The presence of several different aneuploid chromosome numbers within an individual plant is called aneusomaty18. Aneusomaty involves a variation in the number of normal, not B chromosomes. Such variation has been reported in a number of plants including Rubus19 and sugar cane20. In our plants the polysomy of particular chromosomes appeared to increase (Fig 1d, Fig 5). Because the small size of the Paspalum chromosomes made detailed karyotype analysis difficult, confirmation of this result would require meiotic analysis of pollen mother cells (PMC).
A study of Phalaris species and hybrids by Schifino et al.21 showed a wide variation in chromosome number in PMC and root tip mitoses. Mitotic disturbances such as bridges, laggards, multipolar mitoses and micronuclei were reported. In a subsequent paper Schifino and Gus22 showed that this chromosome instability was passed on to seedlings without loss of vigor or fertility. It was suggested that the variation in chromosome number was caused by hybrid genetic imbalance.
Chromosome number instability sometimes arises following hybridisation between polyploid plants 23, 24, 25. A wide variation in chromosome numbers has also been reported in callus cultures 18, 26. Sometimes the chromosome number of plants regenerated from these calli is stable, but in other cases mixoploid plants are produced18, 27.
Chemical or stress induced mitotic disturbance was studied in Allium cepa by Huskins and others in a series of papers28. Multipolar mitoses and micronuclei were associated with variable chromosome numbers in somatic cells. These abnormalities were not observed in the present work.
One possible explanation for the variation observed in the present work is that the chromosomes divided unevenly at mitosis, i.e. mitotic non-disjunction. This would explain the presence of cells which were polysomic for individual chromosomes. Cells with some combinations of chromosomes might divide less frequently and some cell lines might indeed be lost.
Between plant variation
The inheritance of apomixis has been studied in a number of apomictic grasses. In Paspalum notatum apomixis is controlled by a few recessive genes, whereas in Cenchrus ciliaris (=Pennisetum ciliare) apomixis is controlled by a dominant gene which is epistatic to a second gene controlling sexuality29. In C39 there may have been a mutation or masking of a gene promoting apomictic seed production as this somaclone produced very little viable seed. With apomixis being impaired a low level of sexual reproduction may have occurred. The presence of six SC2 seedlings with about 20 extra chromosomes is consistent with sexual activity in the form of fusion of a sperm nucleus (with 20+chromosomes) and an unreduced egg (50 chromosomes). Evidence for fertilization of unreduced eggs in apomictic P.dilatatum has been reported in breeding programmes6, 9. Furthermore, in the faculative apomict Cenchrus ciliaris a small proportion of seeds were shown to result from the fertilization of an unreduced egg(15 of 1252 seeds) or a reduced egg (21/1252)30. These seedlings had 2n=54 and 2n=36 chromosomes respectively, compared with 2n=36 chromosomes in the maternal parent.
The increased chromosome numbers seen in four SC2 plants(C39/1,2,5,10) were inherited by most of their SC3 progeny, and these progeny also retained the maternal phenotype. Moreover, the four SC2 plants invoved showed greatly increased seed viability compared with C39 (Tab 1). This result suggests that these SC2 plants had regained the ability to develop seed apomictically. However, the fact that a few of the SC3 progeny from C39/1,/5 and /10 showed further substantial increases in chromosome numbers suggests that some embryos were still being produced following fertilisation.
In two cases seedlings were produced with a clearly reduced complement of chromosomes compared with their maternal parent. C39/11, an SC2 plant, and C39/2/2, an SC3 plant, had modal chromosome numbers of 48 and 54 respectively. These plants may have resulted from the fusion of sperm nuclei with haploid egg cells.
While there were changes in chromosome numbers in seven SC2 seedlings the other four SC2 seedlings (C39/4, 7-9) had the unchanged 50 chromosome number. Of these latter four plants, three were phenotypically different from C39 which suggest that they were not produced by apomixis and so could have arisen by sexual means.
In conclusion, the varying degrees of aneuploidy in the progeny of C39 indicate that the mode of reproduction in C39 was different from that of Raki. C39 appears to have produced seedlings by a mechanism which involved impairment of apomixis combined with the development of embryos resulting from fusion of sperm nuclei with either reduced or unreduced egg cells. The intra-plant variation in chromosome number (aneusomaty) appears to be an independent phenomenon, resulting from an imbalance of chromosomes in hybrid cells. Our results indicate that further work aimed at determining whether C39 reproduced sexually would be worthwhile and a study of embryo sac development in C39 and Raki is in progress.
References
Burson BL . Phylogenetic investigations of Paspalum dilatatum and related species. In: JA Smith and VW Hays eds, 1983. Proceedings 14th International Grasslands Congress, 1981. Lexington, Kentucky. pp.170–3.
Bashaw EC, Forbes I . Chromosome numbers and microsporogenesis in Dallisgrass, Paspalum dilatatum. Agron J. 1958; 50:441–5.
Bennett HW, Marchbanks WW . Seed drying and viability in dallisgrass. Agron J. 1969; 61:175–7.
Bashaw EC, Voight PE, Burson BI . Breeding challenges in apomictic warm-season grasses. In: JA Smith and VW Hays eds, 1983. Proceedings 14th Internatinal Grassland Congress, 1988. Lexington Kentucky. pp.179–81.
Burton GW . Conventional breeding of dallisgrass, Paspalum dilatatum Poir. Crop Sci. 1962; 2:491–4.
Bennett HW, Burson BL, Bashaw EC . Intraspecific hybridization in dallisgrass, Paspalum dilatatum Poir. Crop Sci. 1969; 9:807–9.
Bashaw EC, Hoff GJ . Effects of irradiation on apomictic common dallisgrass. Crop Sci. 1962; 5:501–4.
Burton GW, Jackson JE . Radiation breeding of apomictic prostrate Dallisgrass, Paspalum dilatatum var. pauciciliatum. Crop Sci. 1962; 2:495–7.
Bennett HW, Bashaw EC . Interspecific hybridization with Paspalum dilatatum Poir. Crop Sci. 1966; 9:807–9.
Larkin PJ, Scrowcroft WR . Somaclonal variation: a novel source of variability from cell culture for plant improvement. Theor.Appl. Genet. 1981; 60:197–214.
Karp A, Bright SWJ . On the causes and origins of somaclonal variation. Oxford survey of plant molecular and cell biology 1985; 2:199–234.
Davies L J, Cohen D . Phenotypic variation in somaclones of Paspalum dilatatum and their seedling offspring. Can J Plant Sci. 1992; 72:773–84
Kao KN . Staining method for protoplasts and cells. In: LR Wetter and F Constabeled eds. Plant Tissue culture Methods (2nd edition). Prairie Regional Laboratory, National Research Council of Canada. 1982.
Karp A, Maddock SE . Chromosome variation in wheat plants regenerated from cultured immature embryos. Theor Appl Genet 1984; 67:249–55.
Evans DA, Sharp WR . Single gene mutation in tomato plants regenerated from tissue culture. Science 1983; 221:949–51.
Hanna WW, Lu C, Vasil IK . Uniformity of plants generated from somatic embryos of Panicum maximum Jacq. (Guinea grass). Theor Appl Genet 1984; 67:155–9.
Swedlund B, Vasil IK . Cytogenetic characterization of embryogenic callus and regenerated plant of Pennisetum americanum (L.) K. Schum. Theor Appl Genet 1985; 69:575–81.
D'Amato F . Cytogenetics of plant cell and tissue cultures and their regenerates. CRC Critical Reviews in Plant Sciences 1985; 3:73–112.
Britton DM, Hull JW . Mitotic instability in Rubus. Journal of Heredity. 1957; 48:11–20.
Heinz DJ, Mee GWR, Nickell LG . Chromosome numbers of some Saccharum species, hybrids and their cell suspension cultures. Amer J Bot. 1969; 56:450–6.
Schifino MT, Zanella CC, Gus R . Chromosome number instability in Phalaris L. Cytologia 1985; 50:89–99.
Schifino MT, Gus R . Transmission of somatic chromosome number instability in Phalaris L. (Gramineae). Rev Brasil Genet 1986; IX, 3:549–54.
Illies ZM . The development of aneuploidy in somatic cells of experimentally produced triploid Larches. Heredity 1966; 21:379–85.
Sadasivaiah RS, Lesins K . Reduced chromosome number in root tip cells of Medicago. Can J Genet Cytol 1974; 16:219–27.
Maliga P, Kiss ZR, Nagy AH, Lazar G . Genetic instability in somatic hybrids of Nicotiana tabacum and Nicotiana knightiana. Molec Gen Genet 1978; 163:145–51.
Bayliss MW . Chromosomal variation in plant tissues in culture. Intern Rev Cytol Suppl 1980; 11a:113–44.
Feher F, Hangyel Tarczy M, Bocsa I, Dudits D . Somaclonal variation in tetraploid alfalfa. Plant Science 1989; 60:91–9.
Huskins CL, Cheng KC . Segregration and reduction in somatic tissue IV. Reductional groupings induced in Allium cepa by low temperature. J Hered. 1950; 41:13–8.
Taliaferro CM, Bashaw EC . Inheritance and control of obligate apomixis in breeding buffelgrass, Pennisetum ciliare. Crop Sci 1966; 6:473–6.
Sherwood RT, Young BA, Bashaw EC . Facultative apomixis in buffelgrass. Crop Sci. 1980; 20:375–9.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhu, J., Davies, L., Cohen, D. et al. Variation in chromosome number in the seedling progeny of a somaclone of Paspalum dilatatum. Cell Res 4, 65–78 (1994). https://doi.org/10.1038/cr.1994.7
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1994.7
Keywords
This article is cited by
-
Taxon delimitation, chromosome numbers and genetic diversity of Paspalum polyphyllum and P. bicilium (Poaceae, Paspaleae)
Plant Systematics and Evolution (2021)