Abstract
Lymphotoxin(LT) is a glycoprotein secreted by activated T cell. The expression of LT gene is mainly regulated at the level of transcription. By using human LT DNA as a probe, we carried out a RNA dot blotting test and found that the longer the time of Jurkat human T-lymphoma cells exposed to the PMA and PHA, the more endogenous LT mRNA could be produced. Results of gel retardation assay showed that the nuclear extract from Jurkat cells treated with PMA and PHA formed different DNA-protein complexes. Changes in complex patterns were observed at various time intervals of PMA and PHA induction. A specific protein-binding site was mapped out to be a 22-bp sequence at the 5′upstream region of human LT gene by DNase I footprinting analysis. This region was similar to the sequence recognized by the proteins of NF- kB family. The results of fragment competition and homology analysis indicated that the 22-bp sequence contains a kB-like motif only, which is located at the base pairs −100 to −90 (5′-GGGGGCTTCCC-3′). Thus, the NF- kB-like factors were involved in the protein-DNA interaction. Furthermore, there were more than one retarded bands appearing in the gel retardation assay. It suggested that there may be several NF- kB-like factors involved in the regulation of LT gene transcription at the same site.
Similar content being viewed by others
Introduction
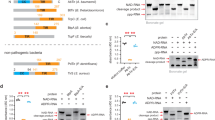
Lymphotoxin(LT), also called tumor necrosis factor β(TNF -β), is a lymphokine released by mitogen or antigen-activated T lymphocytes. Under circumstances of various neoplast and autoimmune diseases, LT production can also be detected respectively, contributing to the cytotoxicity and cytostasis in several immunologic reactions1. In a previous study we have found that there were at least 2 negative regulatory elements(NRE1 and NRE2), located at the 5' flanking sequence of LT gene (Fig 1). In NRE2, there is a motif (spanned from 100 to 90 base pair upstream of LT gene transcription start point) with high homology to kB motif. Deletion in this motif led to CAT expression of reporter vector from a non-responsive state to a stimulative one2. Thus, NF-kB may play an important role in the inducible expression of LT gene.
NF- kB is present in various types of cells and participates in the inducible transcription of diverse cellular genes through binding to various kB motifs. These genes mostly include those involved in host immunological responsiveness in defense3. Although, NF- kB is constitutively present in the nucleus of mature B cells, it behaves as an inducible nuclear factor in other types of cells4.
To elucidate the mechanism involved in the control of LT gene transcription, we have identified a specific binding site for NF- kB-like factors at the 5′ upstream region of human LT gene, by using gel retardation assay and DNase I footprinting analysis. On account of this, experiment designed to purify this kind of nuclear factor(s) and clone the gene(s) for the characterization of their multiple functions in immunological reactions are in progress.
Materials and Methods
Cell lines and cell culture
L929 mouse fibroblasts and Jurkat T-lymphoma cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum(FBS). Stimulation of Jurkat cells was effected by adding both phorbol-12-myristate 13-acetate (PMA, 50 ng /ml) and phytohemagglutinin (PHA, 2 μg/ml).
Nuclear extract preparation
Crude nuclear extracts were prepared by the method of Digman5.
RNA dot blot
RNA dot blotting was performed as described6. The probe was the plasmid pGEM3-huLT , which contains human LT DNA and was constructed in our laboratory 7.
Gel retardation assay
The 333-bp fragment, including the sequence position −305 to +131(−247 to −144 bp deleted) of LT gene, was labeled with [α-32P]dATP. The gel retardation assay was performed as described by Carthew8 with the following modifications: 32P-labeled fragment (2 ng) and 18 μg of nuclear extract were mixed in 75 mM KC l/ 10 mM Tris- HCl(pH7.5)/1 mM dithiothreitol / 1 mM EDTA /1.5 mM Mg Cl2 /3 mM GTP /8 μg BSA /8% glycerol / 1.8 g poly (dI -dC) copolymer , to a final volume of 20 μl. The mixture was incubated for 30 min at room temperature and electrophoresed in a 4% undenatured polyacrylamide gel with a gel running buffer consisting of 6.7 mM Tris-HCl(pH7.5), 3.3 mM sodium acetate and 1 mM EDTA, Gels were directly autoradiographed. For competition experiments, the competitor DNAs were put into the mixture before the addition of labeled fragment.
DNase I footprinting analysis
For DNase I protection, the fragment was labeled at 3′ end of the non-coding strand and the DNA was incubated with nuclear extracts in a above-mentioned binding reaction condition but the volume was scaled up 2.5-fold. After the binding reaction, 5 μl of Ca2+ / Mg2+ solution (10 mM Mg Cl2/5 mM CaCl2) was added and set aside for 1 min at room temperature. The mixture was digested with 2 or 4 U of DNase I(Pharmacia) for 1 min and extracted twice with an equal volume of phenol:chloroform(1:1). The aqueous phase was transferred and the DNA was precipitated with ethanol. Products were subjected to electrophoresis in a 8% polyacrylamide/8 M urea sequencing gel. Labeled fragment DNA, which has been subjected to modified Maxam-Gilbert sequencing reactions7, was electrophoresed to determine the protected sequence.
Homology analysis
Homology among the DNA sequences was analyzed by using the Gene Bank database.
Result
RNA dot blot
Jurkat cells were used to demonstrate the synthesis of LT mRNA induced by PMA and PHA. As shown in Fig 2, L929 cells and unstimulated Jurkat cells did not accumulate a detectable amount of LT mRNA, while PMA and PHA induced an increase in LT mRNA production. During the whole of 30 hours' stimulation, the maximal response for LT mRNA expression occurred at the end of 30 h period. It indicated that Jurkat cells were able to be induced to produce endogenous LT mRNA. In other words, Jurkat cell line is suitable for studying the inducible expression of LT gene, because all of the components essential for transcription regulation, such as system responsed to extracellular stimuli, signal transduction pathways and cis- / trans-acting elements, were functioning in Jurkat cells.
LT mRNA synthesis in Jurkat cells induced by PMA and PHA
Lane 1: DNA of the pGEM-huLT (positive control)
Lane 2: RNA of L929 cells (negative control)
Lane 3: RNA of Jurkat cells untreated
Lanes 4–7: RNA of Jurkat cells exposed to 50 ng / ml PMA and 2 μg /ml PHA for 4 h, 10 h, 20 h and 30 h respectively.
Each RNA dot blotting performed using 2 × 106 cells.
Gel retardation assay
To characterize the protein-DNA interactions with the 5′ flanking sequence of huLT gene, the 333-bp fragment was end-labeled for use in gel retardation assays. Crude nuclear extracts were prepared from Jurkat cells treated with PMA and PHA for 4 h. After incubation of the fragment with the extracts, 4 retarded bands of of nearly equal intensities could be resolved by gel electrophoresis (Fig 3, lane 1). The results yielded 2 important possibilities for further investigation. Firstly, in the crude nuclear extracts of treated Jurkat cells, there may be more than one nuclear factors binding to the site at the 5′ upstream region of huLT gene. Secondly, at the 5′ upstream of LT gene there are several binding sites undertaking the response for the specific protein-DNA interactions. Competition with a 50-fold or more molar excess of fragment B (position −305 to +131 bp, −260 to −128 bp deleted) effectively prevented the formation of complexes represented by band 1, 2 and 4, but not by band 3 (Fig 3, lanes 2-4). It indicated that fragment B contains the same binding site as in the labeled probe. However , even when a 150-fold molar excess of fragment C (position −305 to +131 bp, −272 to −93 bp deleted) was used in the competition reaction, 4 retardation bands still appeared (Fig 3, lane 5). Thus, it means that the sequence specific for protein DNA interaction does not exist in fragment C.
Competition for binding of Jurkat nuclear extracts to the 5′ flanking region of huLT gene
Gel retardation assay of labeled fragment (base pairs −305 to +131, −247 to −144 deleted) was incubated with extracts from Jurkat cells treated with PMA (50 ng/ml) and PHA (2 μg/ml) for 4 h
Lane 1: 15 μg of nuclear extracts in binding reaction
Lanes 2-4: In the presence of 50, 100 and 150-fold molar excess of the competitor fragment B (base pairs −305 to +131, −260 to −128 deleted)
Lane 5: In the presence of 150-fold molar excess of the competitor fragment C (base pairs −305 to +131, −272 to −93 deleted)
DNase I footprinting analysis
To identify the exact sequence involved in protein-DNA interaction, DNase I footprinting analysis was performed as described above. Fig 4 showed that a 22-bp region (5′-GGGCCCGGGGGAAGCCCCCAGGG-3′) with boundaries at positions −104 to −83 was protected by the nuclear extracts from Jurkat cells treated with PMA and PHA for 4 h. (Fig 4, Lanes 3-5). In addition, a footprinting competition test using a 200-fold molar excess of fragment B was carried out. As shown in Fig 4 (Lane 6), the fragment B competed 22-bp region for binding of factors and left this region protein unprotected. These results were consistent with the localization and specificity of the binding site by gel retardation assays.
DNase I footprinting analysis of the 5′ upstream regulatory element of huLT gene
Binding reactions were performed with the labeled fragment (base pairs −303 to +131, −247 to −144 deleted) and nuclear extracts from Jurkat cells exposed to PMA(50 ng/ml) and PHA(2 μg/ml) for 4 h.
Lane 1: G+A reaction of the same fragment used as size markers
Lane 2: No extracts added
Lanes 3–5: 100, 100 or 150 μg of the nuclear extracts and digestion with 2U, 4U or 4U of DNase I for 1 min
Lane 6: 100 μg of extracts and 4U DNase I for 1 min in the presence of 200-fold molar excess of fragment B
Discussion
Investigation of the regulation of gene expression is attracting more and more attention of molecular biologists nowadays. In comparison with prokaryotes, the eukaryotic gene regulation is much more complex. Although an eukaryotic gene may be regulated at several levels, it is generally accepted that initiation of messenger RNA synthesis is the primary control point in the regulation of diverse gene expressions. The interactions of cis- and trans-acting elements play a very important role in the control of transcriptional initiation.
The assay performed in the present study resulted in the identifying a specific protein-binding site located at the 5′ upstream of huLT gene. The multiple complexes formed in gel retardation assays suggested that there existed the site for binding of trans-acting proteins. The difference between the competition tests of fragment B and C can be explained by the results of DNase I footprinting analysis. Fragment B includes the whole 22-bp protected region which is responsible for the specific protein binding. On the other hand, this region is devoid of its 3′ terminal 12-bp in fragment C so that the property of protein binding is no longer discernible.
Our previous study has demonstrated that the integrity of an kB-like motif (position −100 to −90 bp) is essential for the inducible LT gene expression2. This kB- like motif is completely covered by the protected region in the DNase I footprinting analysis. Thus, on the basis of the findings obtained in the present investigation as well as in our earlier experiments2, we concluded that there is only one kB-like motif in the 5′ upstream region of huLT gene acting as a binding site for members of the NF- kB family.
Another question may be put forward: how did one kB-like motif give rise to multiple complexes in the gel retardation assays. Two retarded bands were observed in the incubation with nuclear extracts treated with PMA and PHA for 15 min, but 4 bands when treated for 4 h (Fig 5). This observation suggested that with the transfer from cytoplasm to nucleus, NF- kB-like factors may be modified to molecules of different sizes but still preserve the ability of binding to kB-like motif. As these NF- kB-like factors may be bind to kB-like motif with different affinities, their activities in the competition analysis are different. This may be the reason for the reduced intensity of band 2 in Lane 5 of Fig 3. However, the possibility exists that besides this kind of proteins, additional proteins may contribute to protein-DNA interactions as well. It has been proved that kB-like motif in mouse MHC class I gene is also a binding site for H2TF-1, eg transcription factor of H2 gene9. Therefore, in order to directly characterize the factors involved in this sort of protein-DNA interaction, it is necessary to clone out the gene coding for those factors and to identify the gene products.
Time course for PMA and PHA induction of kB-like site binding proteins
Nuclear extracts from Jurkat cells either untreated (Lane 2) or treated with PMA(50 ng/ml) and PHA(2 μg/ml) for 15 min (Lane 3) or for 4 h(Lane 4) were incubated with the labeled fragment (−305 to +131, −247 to −144 deleted), and the resultant DNA-protein complexes were resolved on 4% polyacrylamide gel. No nuclear extracts add in Lane 1.
References
Kehrl JH, Alvarez-Mon M, Deling GA and Fauci AS . Lymphotoxin is an important T-cell derived factor for human B-cells. Science 1987; 238:1144–6.
Zheng LX, Zhu XH, Li CB and Zhao SY . Functional analysis of the 5′ flanking sequence of human lymphotoxin gene. J Immunol (China), 1990; 6:147–51.
Xu R and Zhao SY . NF -kB and gene regulation. J Immunol (China) (in press).
Leonardo MJ and Baltimore D . NF-k B: A pleiotropic mediator of inducible and tissue-specific gene control. Cell 1989; 58:227–9.
Dignam JD, Lebovitz RM and Roeder RG . Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 1983; 11:1457–78.
Sambrook J, Fritsch EF and Maniatis T . Molecular cloning, Cold Spring Harbor, New York 1989.
Liu JT, Li CB and Zhao SY . Expression of human lymphotoxin gene in CHO-DHFR-cells. Acta Genet sin 1991; 18:208–13.
Carthew RW, Chodosh LA and Sharp PA . An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promotor. Cell 1985; 43:439–48.
Mauxion F and Sen R . Alteration of a single nucleotide allows efficient binding of H2TF1/KBF1 to the immunoglobulin enhancer B motif. Mol Cell Biol 1989; 9:3548–52.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Xu, R., Zhao, S. Identification of an k B-like motif at the 5′ upstream region of human lymphotoxin gene. Cell Res 4, 1–7 (1994). https://doi.org/10.1038/cr.1994.1
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1994.1