Abstract
Defined culture systems supporting spermatogonial differentiation will provide experimental platforms to study spermatogenesis. However, germline-intrinsic signaling mechanisms sufficient to support spermatogonial differentiation without somatic cells remain largely undefined. Here we analyzed EGF superfamily receptor and ligand diversity in rat testis cells and delineated germline-intrinsic signaling via an ERBB3 co-transducer, ERBB2, as essential for retinoic acid-induced syncytial growth by differentiating spermatogonia. Similar to the ERBB2/3 agonist NRG1, we found that KIT Ligand (KITL) robustly supported spermatogonial differentiation without serum or somatic cells. ERBB2 inhibitors failed to disrupt KITL-dependent spermatogonial development, and KITL prevented ERBB3-deficient spermatogonial degeneration upon differentiation. Thus we report that NRG1 and KITL activate alternative pathways downstream of retinoic acid signaling in the germline that are essential for stem cells to undergo premeiotic steps of spermatogenesis in culture. Robust serum/soma-free spermatogonial differentiation opens new doors to study mammalian germ cell biology in culture and to discover factors that can drive meiotic progression in vitro.
Similar content being viewed by others
Introduction
Spermatozoan development is maintained within the testes’ seminiferous epithelium by the process of spermatogenesis.1 Spermatogonial stem cells maintain spermatogenesis by their unique abilities to self-renew or produce spermatogonial syncytia that differentiate through meiosis to form haploid gametes termed round spermatids.2 Round spermatids undergo a complex metamorphosis to develop into fully elongated spermatozoa by the process of spermiogenesis.1 Chemically defined culture systems that support spermatocyte maturation and spermatid development from mammalian spermatogonial stem cell lines remain to be established but, prospectively, will allow researchers to probe diverse premeiotic, meiotic and postmeiotic germ cell processes.1
The ability to study spermatogenesis in vitro is being held at bay because culture systems that robustly support spermatogonial proliferation and/or differentiation into meiosis do not exist for most mammalian species outside rodents. In rodents, donor spermatogonial stem cells can be maintained long term in culture3 but can only be cultured through meiosis in recipient testes4 or in organ culture within seminiferous tubules.5 Going forward, fully defined culture systems that effectively support spermatid production from spermatogonial stem cell lines will need to be established from diverse Animalia to realize the full potential of in vitro spermatogenesis for experimentally dissecting cellular processes and for producing haploid gametes.
‘A-single (As)’ spermatogonia function as spermatogonial stem cells that initiate spermatogenesis during development into syncytia containing 2–32 ‘undifferentiated’ A-paired (Apr) and A-aligned (Aal) progenitor spermatogonia.6,7 In rodents, undifferentiated spermatogonia mitotically arrest during seminiferous epithelial cycle stages VI–VIII and then transform into ‘differentiating’ type A1 spermatogonia under control of KITL and retinoic acid (active vitamin A derivative).8,9 Type A1 spermatogonia re-enter the mitotic cell cycle and give rise to subsequent generations of differentiating spermatogonia (types A2>A3>A4>Int>B),10 by which time germ cell numbers/syncytium can be amplified >100-fold prior to entering meiosis to form spermatocytes.11
Polypeptides encoded by glial cell line derived neurotrophic factor (Gdnf) and fibroblast growth factor (Fgf) are essential for mouse spermatogonial stem cells to maintain spermatogenesis.12,13 GDNF and FGF2 also support proliferation of mouse,14,15 rat16,17 and hamster18 spermatogonial stem cells in culture. In fact, GDNF and FGF2 are essential components in serum-free media that maintain mouse spermatogonial stem cell proliferation long term without somatic cells.19 WNT-family polypeptides accelerated mouse spermatogonial stem cell proliferation in a serum-free medium containing GDNF, GFRa1 and FGF2 but did so indirectly by signaling in early progenitor spermatogonia.20,21 In contrast to highly defined culture systems that maintain spermatogonial stem cell proliferation, a large knowledge gap exists on signaling pathways that can act directly in germ cells to support additional premeiotic, meiotic or postmeiotic spermatogenic differentiation steps independent of a somatic environment.
KITL is a prominent Sertoli cell-derived polypeptide growth factor essential for differentiating spermatogonia development in vivo.8 Signaling via the KITL receptor KIT is also essential for differentiating spermatogonia development in vivo.22,23 KIT has been reported on differentiating spermatogonia, early spermatocytes and Leydig cells.24,25 Retinoic acid is a vitamin A-derived steroid hormone and is known for its ability to regulate spermatogenic cell differentiation in testes,26 organ cultures,27,28 isolated testis cell cultures,29 cultures enriched with prospermatogonia30 and mouse spermatogonial lines.31 Still, to date, neither KITL nor retinoic acid have been defined as essential to support robust clonal development/syncytial growth of differentiating spermatogenic cells without somatic cells.
One germline receptor, ERBB3, was recently genetically annotated as essential for syncytial growth of differentiating spermatogenic cells in a serum/soma-free medium containing NRG1, GDNF, FGF2 and retinoic acid (i.e., SD Medium).32 However, Erbb3-null germlines fully supported spermatogenesis in recipient rat testes, indicating that additional spermatogonial growth factors existed in rats.32 Moreover, NRG1 is an ERBB3-ligand, but Sertoli cell-derived Nrg1 was specifically required for meiosis in mice.33 Here, by in-depth analysis of EGF-family signaling molecules expressed in rat spermatogenic cells and growth factor components in SD Medium, we have defined alternate ERBB2-dependent and ERBB2-independent growth factor signaling pathways that act directly in the rat germline with retinoic acid to robustly support syncytial growth of differentiating spermatogonia without somatic cells.
Results
ERBB2 and ERBB3 are selectively detected in rat spermatogonia
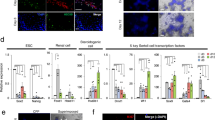
ERBB3 (HER3 in humans) is encoded by one of four different mammalian erythoblastoma virus B homolog genes (i.e., Erbb1, Erbb2, Erbb3, Erbb4) (Figure 1a).34 In somatic tissues, ligand-bound ERBB3 signals as an obligate heteromer with other ERBB-family transmembrane receptor tyrosine kinases.34 To identify potential signaling partners for ERBB3 in the germline, we analyzed ERBB-family proteins in isolated rat spermatogenic and somatic testis cells by western blotting. ERBB2 and ERBB3 (~185 kDa bands) were detected in freshly isolated undifferentiated type A spermatogonia and in a rat spermatogonial stem cell line enriched for the GDNF/GFRa1 signal transducer, RET (Figure 1b).
ERBB-family signaling molecules in rat testis cells. (a) Polypeptides in the EGF super-family signal by activating ERBB-family transmembrane receptor tyrosine kinases. ERBB1 is a receptor for ‘classical’ low molecular weight EGF-like peptides. ERBB2 is the primary transducer for ligand-bound ERBB1, ERBB3 and ERBB4. ERBB2’s extracellular domain does not bind known ligands. ERBB3 is a receptor for Neuregulin-1 (NRG1), NRG2 and Neuroglycan-C (CSPG5). Ligand bound ERBB3 displays poor kinase activity and signals most effectively as a heteromer with ERBB1, ERBB2 and/or ERBB4. ERBB4 is a receptor for NRG1, NRG2, NRG3 and NRG4 plus other EGF-like peptides*. (b) Western blotting analysis of ERBB-family proteins in fractions of testis cells from 23-day-old rats. Lysates of type A spermatogonia after proliferating for ~180 days/15 passages in culture (SgL), freshly isolated laminin-binding type A spermatogonia (Sg), laminin non-binding spermatogenic cells (Scy), tubular somatic cells (SC), interstitial somatic cells (IC), MCF7 human mammary gland cells (MCF) and COS7 monkey kidney cells (COS). Arrowheads: ERBBs 1–4 (~185 kDa), RET (~155 and 170 kDa) and TUBA1a (~55 kDa). (c) Relative abundance (qtPCR) of ERBB-family transcripts in testis cells isolated from 23-day-old rats (n=cells from three different rats; ±S.E.M.). Spermatogonia (Sg), Spermatocytes (Scy; differentiating spermatogonia/early spermatocytes), Tubular somatic cells (SC) and Interstitial somatic cells (IC) are cell types described in panel (b). (d) Testis cross-section from 26-day-old tgGCS-EGFP transgenic rats labeled with anti-ERBB2 (Red) overlaying EGFP fluorescence from germ cells (green). Note, cytoplasmic ERBB2 labeling in germ cells resembling differentiating spermatogonia (white arrows) and spermatocytes (yellow arrow). Scale, 40 μm. (e) Rat seminiferous tubule whole mount from 24-day-old wild-type rat labeled using antibodies to ERBB2 (Red) and ZBTB16 (Green). Scale, 20 μm. Note: nuclear ZBTB16 labeling is more robust in ERBB2-dim spermatogonia (cyan arrows), compared with ERBB2-bright spermatogenic cells (white arrows). (f) Rat seminiferous tubule whole mount from a 24-day-old wild-type rat labeled with antibodies to ERBB2 (Red) and phospho-Histone-3 (pH3, Green). Scale, 40 μm. Note: nuclear pH3 in large mitotic ERBB2+ syncytia.
ERBB3 was detected at similar levels in differentiating spermatogonia/early spermatocyte preparations compared with the less differentiated type A spermatogonia (Figure 1b). However, ERBB2 abundance was reduced in differentiating spermatogonia/early spermatocytes relative to type A spermatogonia (Figure 1b). Compared with ERBB2 and ERBB3, ERBB1 and ERBB4 were most abundant in interstitial and tubular somatic cells, respectively (Figure 1b). A relatively weak ERBB4 band was detected in freshly isolated type A spermatogonia, but not in the rat spermatogonial line (Figure 1b). Erbb-family transcript profiles generated by qtPCR and RT-PCR (Figure 1c, Supplementary Figure S1a) correlated with the relative abundance of ERBB-family proteins in testis cells by western blotting (Figure 1b). Thus ERBB2 and ERBB3 were the predominant ERBB-family members detected within rat spermatogenic cells.
We previously detected ERBB3 by antibody labeling selectively on a novel sub-population of type A single spermatogonia in rats.35 Because ERBB2 was detected as a potential signaling partner for ERBB3 in spermatogonia (Figure 1b), we analyzed ERBB2 localization in sections and whole mounts of rat seminiferous tubules. ERBB2 labeling was detected on spermatogenic cells (tgGCS-EGFP+) (Figure 1d) and selectively in differentiating type A spermatogonia (Figure 1e; white arrows, ZBTB16lo). ERBB2 localized to spermatogonial cytoplasm and was enriched at the plasma membrane of larger, synchronously dividing syncytia characteristic of differentiating spermatogonia (Figure 1f). Weaker ERBB2 labeling intensities were detected in spermatocytes (Figure 1d; yellow arrow) and undifferentiated type A spermatogonia (Figure 1e; cyan arrows; ZBTB16hi). Detection of ERBB2 on differentiating type A spermatogonia provided a candidate co-transducer for ERBB3-dependent development of differentiating rat spermatogenic cells in serum/soma-free cultures.32
Rat testes express an array of EGF-family ligand mRNAs
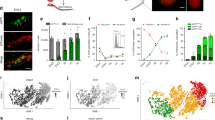
To analyze potential ligands for ERBB-family receptors in testes, we first surveyed transcripts encoding NRG1 variants expressed by germline and somatic testis cells (Figure 2a). Nrg1 was selectively detected in rat type A spermatogonia by RT-PCR (Supplementary Figure S1b). Full open reading frames encoding secreted (Type 1β3) and transmembrane (Types 1β2a and 1α2b) Nrg1 mRNA variants were cloned from type A spermatogonia (Figure 2b, Supplementary Figure S1c).
Spermatogenic cells selectively express neuregulin-family genes. (a) Strategy to analyze Neuregulin-1 (Nrg1) mRNA splice variants. Exons (boxes) in Types I, II and III Nrg1. Distinct NRG1 Types are generated by alternatively splicing N-terminal exons (NDF, Kringle and CRD domains). IgG and Sp exons encode heavily glycosylated domains, which bind heparin sulfate proteoglycans.58 EGF homology domains (black boxes) together with either α or β domains (gray boxes) bind with high affinity to ERBB3 and ERBB4 extracellular domains. Exons designated by tan boxes 1–4 encode extracellular ‘stalk domains’, immediately upstream from the transmembrane domain (TM). Stalk domains 1, 2 and 4 are substrates for distinct metalloproteases, which regulate NRG1 extracellular domain shedding.59 NRG1’s with stalk domain 3 contain a C-terminal stop codon before the TM and are secreted. Exons in the gray boxes encoding different cytoplasmic domains that also regulate extracellular domain shedding. Arrows: PCR primers used to analyze testicular Nrg1 variants. (b) Full-length Nrg1 transcripts amplified from undifferentiated type A spermatogonia encode variants of Type I, NRG1. Arrows: respective PCR primers used to clone Nrg1 variants. (c) Spermatogonia selectively express mRNAs encoding NRG1 and CSPG5 (qtPCR), n=cells from three different rats; ±S.E.M. Spermatogonia, Spermatocytes (differentiating spermatogonia/early spermatocytes), Tubular somatic cells and Interstitial somatic cells refer to Sg, Scy, IC and SC described in Figure 1b. (d) Western blots for NRG1α1 and NRG1β1 in the rat brain (B) and testis (T) and a primary spermatogonial line derived from undifferentiated type A spermatogonia (Sg). Arrows on left of each blot represent respective size molecular markers (kDa). Blue asterisks denote respective size NRG1 variants previously reported in the rat brain.36–38 (e) Immunolabeling for NRG1α1 (red cytoplasm) in adult rat testis sections. Nuclei counterstained with Hoechst 33342 dye (cyan). Pl=preleptotene spermatocyte; L=leptotene spermatocyte; eZ=early zygotene spermatocyte; P=mid-to-late pachytene spermatocytes; S8, S9, S12, S19=Step 8, 9, 12, 19 spermatids; VE=vascular endothelial cell; L=Leydig cell; S=Sertoli cell. Roman numerals denote spermatogenic stages. Scale bar, 100 μm.
Similar to Nrg1, transcripts encoding other neuregulins, including Ngr2, Ngr3 and Csgp5 were selectively detected in spermatogenic cells (Figure 2c, Supplementary Figure S1b). Nrg1 and Csgp5 were most abundant in type A spermatogonia (Figure 2c, Supplementary Figure S1b), whereas Nrg3 was most abundant in differentiating spermatogonia/early spermatocyte fractions (Figure 2c, Supplementary Figure S1b). In addition to transcripts encoding Neuregulins, spermatogenic cells expressed transcripts encoding multiple ligands for ERBB1 (Egf, Tgfa, Areg, Btc) and ERBB4 (Btc, Hbefg) (Supplementary Figure S1b).
Western blotting for ERBB3 agonists in rat testis and undifferentiated type A spermatogonia identified NRG1α1 and NRG1β1 variants/isoforms previously reported in the rat brain (Figure 2d, cyan asterisks).36–38 NRG1α1 and NRG1β1 ~65 kDa forms were most abundant in total testis; however, NRG1β1 ~75 and ~50 kDa forms were enriched in undifferentiated spermatogonia (Figure 2d). Antibody labeling for NRG1α1 and NRG1β1 variants in adult rat testis sections clearly detected cytoplasmic NRG1α1 in spermatogenic cells ranging from mid-late pachytene spermatocytes to step 17 elongating spermatids and also in Leydig cells (Figure 2e, Supplementary Figure S1d). NRG1α1 labeling was near background levels in spermatogonia, pre-leptotene to early pachytene spermatocytes and Sertoli cells (Figure 2e, Supplementary Figure S1d). NRG1β1 labeling was the strongest in late pachytene spermatocyte cytoplasm and Sertoli cell cytoplasm but less intense to background levels in other testis cell types (Supplementary Figure S1d). Thus rat testis cells collectively expressed transcripts encoding diverse EGF-family ligands and receptors, whereas rat type A spermatogonia selectively expressed gene products linked to activation of ERBB2 as a signal transducer for NRG1 and ERBB3 (Figure 2, Supplementary Figures S1b–d).
ERBB2 inhibitors disrupt spermatogonial differentiation in a serum-free medium
Because ERBB2 was identified as a candidate NRG1/ERBB3 co-transducer in spermatogonia (Figures 1b–f), we tested the ability of highly selective ERBB2 inhibitors to block clonal development of differentiating spermatogenic cells in vitro on feeder layers of mouse embryonic fibroblasts (MEFs) (Figure 3). Rat spermatogonial stem cells were passaged and maintained for 1 week in spermatogonial culture medium (SG Medium) containing FGF2 but without GDNF (SGF Medium; Table 1) to increase the percentage of As and Apr spermatogonia in cultures. To promote differentiation, cultures were shifted to the same medium containing all-trans-retinoic acid (ATRA), which formulated SDFA Medium (Table 1). Spermatogonia were allowed to differentiate in SDFA Medium for 6 additional days prior to analysis. Three ERBB2 inhibitor types were tested with and without ATRA: (1) the small molecule Lapatinib, (2) an ERBB2-neutralizing antibody, and (3) soluble extracellular ERBB3/ERBB4 domains (to deplete NRG-family ligands). Each inhibitor modality effectively and selectively disrupted clonal development of differentiating spermatogonial syncytia (SALL4−, DAZL+) on MEFs in SDFA Medium (Figures 3a and b; Supplementary Figure S2).
ERBB2 inhibitors block syncytial growth of spermatogenic cells. (a) ERBB2 inhibitors selectively block development of rat spermatogonia syncytia on MEFs in response to 3 μM all-trans retinoic acid (ATRA). Lapatinib (1 μM), Fc (6 μg/ml), FcERBB3+FcERBB4 (3 μg/ml+3 μg/ml), Goat IgG (6 μg/ml) and Goat ERBB2-neutralizing IgG (6 μg/ml). Representative of duplicate colony forming assays (±S.E.M., triplicate wells); culture conditions described in panel (b). Scale, 100 μm. (b) Rat spermatogonial line maintained on MEFs in SGF Medium for 1 week and then for 6 additional days in SGF Medium containing ATRA and/or Lapatinib, as in panel (a). Cells were labeled with antibodies to DAZL (Red) and SALL4 (Green) prior to scoring spermatogenic units. (c) Left, ERBB2 inhibition does not attenuate EDU incorporation by spermatogonia. Right, Spermatogonial cultures were passaged into SGF Medium and maintained for 48 h before treating with retinoic acid for 4 days and then pulsing with EDU (red) for 7.5 h pior to fixing and antibody labeling for DAZL (green). Scale, 100 μm. *Note: shorter incubation times in SGF and SD media enabled effects of Lapatinib on EDU incorporation to be captured in longer syncitia prior to their degeneration upon differentiation in response to ATRA. (d) Images of a rat spermatogonial line after culture on MEFs in SGF Medium for 7 days (D1–D8) and then in SGF Medium supplemented with either GDNF or ATRA for 8 days (D8-D16). Note: Lapatinib selectively blocked survival of differentiating spermatogonia in response to ATRA. Scale, 50 μm. (e) (Top) Diagram of culture experiment shown in panel (a), prior to transplanting respective spermatogonial cultures into recipient rat testes (n=4–6 rats/condition with right testis transplanted; P-values, multiple t-tests). (Bottom) Relative numbers of spermatogenic colonies generated/testis/1000 donor germ cells (tgGCS-EGFP+) harvested from cultures described in panel (d). Recipients analyzed 28 days posttransplantation.
Lapatinib treatment did not affect relative numbers of As, Apr and Aal spermatogonia that incorporated 5-ethynyl-2’-dexoyuridine (EDU) (7.5-h pulse) into newly synthesized DNA after 4 days in SDFA Medium on MEFs (Figure 3c). It is important to note that a shorter preincubation time in SGF Medium and a shorter incubation time in SDFA Medium was applied to better capture viable spermatogonial syncytia incorporating EDU in the presence of Lapatinib (Figure 3c). Lapatinib also did not influence relative numbers of functional spermatogonial stem cells maintained in MEFs under basal conditions without ATRA (Figures 3d and e). Lapatinib did reduce a GDNF-dependent increase in stem cell numbers (~twofold) back to basal levels in the absence of ATRA (Figure 3e). Differentiating spermatogenic cells isolated following treatment with ATRA, either with or without Lapatinib, were effectively depleted of spermatogonial stem cells (Figure 3e). Because Lapatinib did not change rates of spermatogonial development into syncytia based on EDU incorporation (Figure 3c) and did not change basal spermatogonial stem cell numbers/culture (Figure 3e), an ERBB2-dependent mechanism was identified that selectively regulated survival of differentiating spermatogonia.
ERBB2 signals soma-free survival of differentiating spermatogonia
Next, we tested spermatogonial growth in SGF Medium supplemented with distinct combinations of GDNF, NRG1 and ATRA on a laminin matrix (Figure 4a). Under each condition tested, ATRA did not sustain development of spermatogonial syncytia without NRG1 (Figure 4a). However, with NRG1, ATRA effectively promoted syncytial growth (Figure 4a). The combination of FGF2, GDNF, NRG1 and ATRA (SD Medium; Table 1) was most effective at supporting accumulation of spermatogonial syncytia (Aal), as well as increasing their relative numbers over that of As spermatogonia (Figure 4a). Spermatogonial development in SD Medium on laminin appeared similar to SGF Medium supplemented with ATRA and MEFs (Figures 3a and b).
NRG1/ERBB3/ERBB2-dependent growth of differentiating spermatogonia. (a) ATRA and Neuregulin-1 stimulate development of aligned spermatogonia. tgGCS-EGFP+ spermatogonial scored/well after 1 week of culture in SGF Medium (Table 1) on laminin and then 6 additional days with and without ATRA (3 μM) in combination with GDNF (0.1 nM) and/or NRG1 (5 nM). Representative of duplicate in vitro spermatogenesis colony-forming assays scored after 2 days in SGF Medium and 6 days in SD Medium (±S.E.M., triplicate wells). (b) Effects of various ERBB-family receptor agonists on soma-free rat spermatogonial cultures on laminin. Respective ERBB agonists were substituted for NRG1 in SD Medium. Representative of duplicate in vitro spermatogenesis colony forming assays (±S.E.M., triplicate wells). Inset: western blotting for pAKT-S473 (arrowhead) and p42/44 (lower bands) in spermatogonia in SGF Medium for 2 days, prior to culturing with and without ATRA for 4 days, and then treating with and without NRG1 for 20 min before lysing. (c) ERBB2 inhibitors disrupt NRG1-dependent spermatogonial development on laminin. Representative of duplicate colony-forming assays scored (±S.E.M., triplicate wells). (d) Images of NRG1-dependent syncytial growth of tgGCS-EGFP+ spermatogenic cells on laminin. Scale, 100 μm. Nuclei labeled with Hoechst 33342 dye.
To test additional ERBB-family receptors for spermatogenic effects, we compared the ability of highly selective ERBB1, ERBB3 and/or ERBB4 agonists to support soma-independent spermatogonial differentiation on laminin in SD Medium. Consistent with effects of knocking out Erbb3 in the germline32 and detection of ERBB2 and ERBB3 in spermatogonia (Figure 1b), only the ERBB3 agonist NRG1 stimulated clonal development of differentiating spermatogenic cells in SD Medium on laminin (Figure 4b). Several other polypeptide agonists that selectively activate ERBB1 and/or ERBB4 were ineffective compared with NRG1 (Figure 4b).
Accordingly, ATRA upregulated the ability of NRG1 to stimulate phosphorylation of the survival factor AKT1 (Serine 473) in differentiating spermatogonia (Figure 4b; inset). And, consistent with spermatogonial cultures on MEFs, NRG1-dependent development of differentiating spermatogonia on laminin was effectively blocked by Lapatinib or the ERBB2-neutralizing antibody (Figure 4c). Because the tgGCS-EGFP transgene is a viable germ cell marker,39 it became clear by co-labeling nuclei with Hoechst 33342 dye in the absence of feeder cells that NRG1 was required for survival of differentiating spermatogonia on laminin (Figure 4d). Thus ERBB agonist selectivity (Figure 4b), ERBB2 inhibitor sensitivity (Figure 4c) and NRG1-dependent AKT-Ser473 phosphorylation (Figure 4b) delineated a NRG1/ERBB3/ERBB2 signaling pathway in SD Medium that supported soma-independent survival of differentiating spermatogonia.
ERBB2/3 and KIT regulate alternate spermatogenic pathways in vitro
Despite an inability to support spermatogonial differentiation in culture, Erbb3-null germlines robustly supported spermatogenesis in recipient testes,32 which indicated that spermatogonial cultures lacked spermatogenesis growth factors present in testes. Thus we sought an alternate pathway that could rescue spermatogenic development on the Erbb3-null background in SD Medium.
Because KIT is expressed on differentiating spermatogonia,24,25,40 and KIT was required for spermatogonial differentiation in vivo,22,41,42 KITL provided a top candidate growth factor to rescue Erbb3-null germline defects. Moreover, KITL elicited antiapoptotic effects on differentiating spermatogonia, spermatocytes and spermatids in rat seminiferous tubule cultures.43 Sterilizing Kit mutations have long been linked to the dominant white-spotting (W) locus in mammals.23 Yet, by unknown mechanisms, mouse spermatogonial lines harboring sterilizing Wv/Wv Kit mutations supported spermatogenesis in recipient testes on a W/Wv C57BL/6 genetic background.44
Indeed, here KITL effectively supported retinoic acid-induced spermatogonial differentiation (DAZL+, TEX14+, PLZF−) on laminin (Figure 5a). The combination of NRG1 and KITL promoted formation of large spermatogenic syncytia containing >32 cells after the 6-day culture period (Supplementary Figure S3). KITL-dependent spermatogonial development was not blocked by ERBB2 inhibitors (Figure 5b), and KITL fully rescued spermatogonial differentiation on the Erbb3-null background (Figure 5c). Moreover, NRG1- and/or KITL-dependent spermatogonial growth in SD Medium effectively depleted spermatogonial stem cells from cultures (Figure 6). Thus NRG1 and KITL were each essential for soma-free, clonal development of differentiating spermatogenic cells in SD Medium.
KITL supports spermatogonial differentiation independent of ERBB2. (a) Images of NRG1- and KITL-dependent spermatogonial differentiation after 2 days in SGF Medium and 6 days in SD Medium. Spermatogonia are labeled with the germ cell marker DAZL (green); nuclei labeled with Hoechst 33342 dye. Scale, 100 μm. (b) Spermatogonia cultured as in panel (a) labeled with antibodies to the germ cell marker TEX14 (green) and a marker for undifferentiated type A spermatogonia, ZBTB16 (red); nuclei labeled with Hoechst 33342 dye. Scale, 50 μm. (c) ERBB2 inhibitors selectively disrupt NRG1-dependent but not KITL-dependent spermatogonial development on laminin. Representative of duplicate colony-forming assays (±S.E.M., triplicate wells). (d) KITL rescues spermatogonial syncytial growth of Erbb3-deficient germlines on laminin. Representative of duplicate colony-forming assays (±S.E.M., triplicate wells).
Spermatogonia cultured in SD Medium lose germline stem cell activity. (a) Diagram of spermatogonial culture experiment on laminin matrix prior to transplanting respective spermatogonial cultures into recipient rat testes (n=5 rats/condition with right testis transplanted; P<0.0001 control versus each test condition, multiple t-tests). (b) Left: Relative numbers of spermatogenic colonies generated/testis/1000 donor germ cells (tgGCS-EGFP+) harvested from cultures described in panel (a) after 1 week in respective SD Media containing NRG1 and/or KITL (D4–D11). Donor ‘Control’ cultures were maintained on laminin in SG Medium the full 10 days (D1–D11) prior to transplantation. Recipients analyzed 28 days posttransplantation. Right: Images of recipient rat testes after being transplanted with spermatogonia from each respective culture condition. Scale, 1 cm.
Discussion
Lack of success at culturing mammalian stem cells through spermatogenesis in defined systems has fueled hypotheses that the complexity of specific germ cell and somatic cell relationships in testes is essential for the process spermatogenesis, which explains why highly pure mammalian spermatogonia have not developed into meiosis, much less through meiosis without somatic cells. Here, we demonstrate that despite intricate germ cell and somatic cell associations in the seminiferous epithelium,1 it remains possible to molecularly dissect actions of spermatogenic growth factors in the ‘test tube’ following differentiation of primary spermatogonial stem cell lines. Systematic addition of candidate factors to differentiating spermatogonial clones in culture without somatic cells provides a strategy to derive haploid gametes in vitro.
By combining pharmacological and genetic approaches to study a defined spermatogonial culture system, we delineated a NRG1/ERBB3/ERBB2 signaling pathway in the germline that supported premeiotic steps of spermatogenesis in vitro (Figure 4). Once ERBB-family subunits that transduced NRG1 signaling in spermatogonia were elucidated in a defined system, we were able to test our hypothesis that an alternate, ERBB-independent pathway could support spermatogonial development in vitro without somatic cells. Based on studies where testicular KIT activity was required for spermatogonial development,22,23,42 we demonstrated that a primary candidate, KITL, could in fact stimulate soma-free syncytial growth of differentiating spermatogonia, which phenocopied NRG1-dependent effects on germ cell development in vitro (Figure 7).
Soma-independent development of differentiating spermatogonia. FGF2 and GDNF support spermatogonial stem cell renewal and syncytial growth of early undifferentiated spermatogonial progenitors (DAZL+, TEX14+, ZBTB16+) in culture on laminin. Retinoic acid acts in the germline to drive transformation of undifferentiated spermatogonia into nascent differentiating spermatogonia. Polypeptide growth factors NRG1 and KITL are required for survival of differentiating spermatogonia on laminin without somatic cells. In the absence of NRG1 and KITL, ATRA drives undifferentiated spermatogonia to undergo development into syncytia of ~4-16 DAZL+, TEX14+, ZBTB16− spermatogenic cells that degenerate.
In mouse testes, ATRA rapidly signaled KIT synthesis by directing repressed Kit mRNAs into polysomes.45 In mouse undifferentiated spermatogonia cultured on fibroblasts, ATRA upregulated Kit and promoted Kit translation by repressing germline miRNAs 221/222.46 Moreover, in a separate study, miRNA 146 was found to block accumulation of Kit transcripts/KIT protein in undifferentiated spermatogonia on fibroblasts, and ATRA aptly downregulated miRNA 146.47 Here we found that most NRG1-like activity supporting ATRA-induced spermatogonial differentiation on fibroblasts was ERBB2 dependent (Figures 3a and b). Thus ERBB2 was either acting downstream of KITL or via an independent pathway in spermatogonia, with the later scenario being the case (Figure 5).
Control to dissect cellular processes using a defined culture system was exemplified by NRG1’s mechanism of action on spermatogonial development reported here, as compared with NRG1’s effects on spermatogonial development using a complex culture system.48 NRG1 was purified from MEF conditioned medium by its ability to synergistically stimulate syncytial growth of differentiating spermatogonia with GDNF and serum.48 In serum-containing medium, NRG1’s effectiveness on spermatogonial syncytia growth was fully dependent on GDNF,48 whereas here NRG1 effectively stimulated spermatogonial syncytial growth independent of GDNF but in combination with ATRA and FGF2 (Figure 4a). Thus it should be determined whether serum factors biologically distinct from ATRA can drive spermatogonial differentiation in culture with NRG1 and GDNF.48
Robust spermatogonial differentiation from stem spermatogonia under highly controlled culture conditions provides a platform to identify factors that can stimulate germ cells to undergo meiosis in vitro. Our studies highlight how alternate signaling pathways regulated by NRG1 and KITL can support spermatogonial development in vitro (Figure 7), which conceivably contribute to the genetic robustness of spermatogenesis in vivo.32,44 Existence of alternate growth factor pathways that act directly in spermatocytes will prospectively facilitate discovery of additional factors that cooperate with NRGs, KITL and/or retinoic acid to promote meiotic progression from differentiating spermatogonia in the culture dish.
Materials and Methods
Animal care and use
Protocols for the use of rats in this study were approved by the Institutional Animal Care and Use Committee at UT-Southwestern Medical Center in Dallas, TX, USA as certified by the Association for Assessment and Accreditation of Laboratory Animal Care International. Rats used for this study were housed in individually ventilated, Lab Products 2100 cages (Seaford, DE, USA) in a dedicated room with atmosphere controls set to 72 °F, 45–50% humidity during a 12-h light/dark cycle (i.e., light cycle=0600–1800 hours, Central Standard Time adjusted for daylight savings time). Rats were fed Teklad LM-485 mouse/rat diet, 5% fat diet (cat no. 7912, Harlan, Inc., Indianapolis, IN, USA) and a continuous supply of reverse osmosis water. Wild-type Sprague Dawley rats were from Harlan, Inc. (Houston, TX, USA) and Brown Norway rats were form Charles Rivers, Inc. (Kingston, NY, USA).
Spermatogonial culture
Rat spermatogonial lines were derived from freshly isolated laminin-binding (LB) spermatogonia and maintained using spermatogonial culture medium (SG Medium), as previously described.49,50 The base SG Medium formulation contained: Dulbecco’s Modified Eagle’s Medium:Ham’s F12 Nutrient Mixture (1 : 1), 6 ng/ml GDNF, 6 ng/ml FGF2, 100 μM 2-mercaptoethanol, 6 mM L-glutamine, and a 1x concentration of the B27-Minus Vitamin-A Supplement solution. Spermatogonia were sub-cultured on feeder layers of MEFs.49,50 Prior to in vitro or in vivo spermatogenesis colony-forming assays, spermatogonia were harvested from MEFs and incubated for 3–4 h on gelatin-coated plates in SG Medium to remove feeder cells.
Isolating undifferentiated type A spermatogonia
Seminiferous tubules were isolated from testes of 23–24-day-old homozygous SD-Tg(ROSA-EGFP)2-4Reh or WT Brown Norway (Charles Rivers, Inc.) rats. The most advanced germ cell types in 23-day-old male rats are spermatogonia and spermatocytes.51 Rats of the SD-Tg(ROSA-EGFP)2-4Reh line were produced by pronuclear injection and are referred to as GCS-EGFP rats, because they exhibit germ cell-specific expression of enhanced green fluorescent protein (EGFP).39 Tubules were enzymatically and mechanically dissociated into a cellular suspension to generate cultures of testis cells in serum-containing medium.52,53 The testis cell cultures were then used to isolate enriched populations of LB spermatogonia by established methods52,53 that first remove >95% of somatic testis cells from the germ cell population by negative selection on plastic and collagen, before positive selection for the spermatogonial stem cells based on their ability to bind to laminin.53 By this procedure, freshly isolated LB germ cells contain >90% undifferentiated, type A spermatogonia (PLZF+, DAZL+) in the single (~88%) or paired (~12%) cell state.48,54 Note, fractions of LB spermatogonia isolated by this procedure contain ~4% somatic cells and ~5% differentiating spermatogonia/early spermatocytes; and fractions of laminin-non-binding spermatocytes were >95% differentiating spermatogonia/early spermatocytes.54
In vitro spermatogenesis colony-forming assay
Spermatogenic colonies formed in vitro were scored after plating ~1.5×103 spermatogonia/well (0.96 cm2) in spermatogonial differentiation medium (SD Medium) at 36.5 °C, 5% CO2. Wells were precoated with ~5 μg/cm2 laminin as described.49 SD Medium is a modified formulation of the serum-free, SG Medium.50 To make SD Medium, SG Medium was modified to contain 6 ng/ml FGF2 (PGF0023, Life Technologies, Inc., Grand Island, NY, USA), 2 ng/ml GDNF (512-GF, R&D Systems, Inc., Minneapolis, MN, USA), 40 ng/ml NRG1β1 T176-K246 (396-HB, R&D Systems, Inc.) and 3 μM ATRA (R2625, Sigma, Inc., St. Louis, MO, USA). KITL (455-MC-010, R&D Systems, Inc.) was supplemented with SD Medium at 100 ng/ml with or without NRG1 in respective experiments.
Under standard, soma-free bioassay conditions using laminin matrix, on ‘day 1’ (d1) spermatogonia were seeded in SGF Medium (without GDNF; Supplementary Table S1) and, except where noted, were maintained for 48 h (n=3 wells/line) before shifting cultures into SD Medium on d3. Medium was changed again on d5 and d7 using fresh SD Medium. On d9 (i.e., 6 days in SD Medium), medium was removed and cells were fixed in 4% paraformaldehyde, sodium phosphate buffer (0.1 M, pH 7.2) for 20–30 min on ice. Spermatogenic units were scored/well (n=3 wells/line/experiment, ±S.E.M.) by counting numbers of ‘Single’, ‘Paired’ and ‘Aligned’ spermatogenic cells. One ‘Aligned’ Spermatogenic Unit=1 syncytium of tgGCS-EGFP+ spermatogenic cells containing 4–32 Hoechst 33342-labeled nuclei. The terms ‘Aligned’ or ‘Aal’ were based purely on morphology and thus inclusive of ‘undifferentiated’ (ZBTB14+ or SALL4+) and ‘differentiating’ (ZBTB14− or SALL4−) spermatogonia in graphs.
Cells were postfixed in methanol for 5 min on ice and then washed 2× with phosphate-buffered saline (PBS) prior to fluorescence labeling with Hoechst 33342 dye and/or antibodies to ZBTB14 (0.2 μg/ml, Clone 2A9, 39987, Active Motif, Inc., Carlsbad, CA, USA), SALL4 (0.2 μg/ml, H0005716-M03, clone 6E3; Abnova, Taipei City, Taiwan), DAZL ~5 nM53 and/or TEX14 ~10 nM.54 Spermatogenic colony scoring and imaging was conducted using an IX70, Olympus fluorescence microscope (Olympus Inc., Waltham, MA, USA) equipped with the Simple-PCI software (C-Imaging Systems, Compix, Cranberry Township, PA, USA). As quality control, representative tgGCS-EGFP+ spermatogonial lines were verified as being germ cells by independently labeling with DAZL IgG and TEX14 IgG prior to (d3) and following differentiation in SD Medium (d9). For experiments utilizing MEFs in Figure 3 and Supplementary Figure S2, spermatogonia were preincubated in SGF Medium on MEFs for 6–8 days, as indicated in legends. Longer preincubation times in SGF Medium increased the percentage of As spermatogonia in cultures. Lapatinib (L-4804) was from LC Laboratories (Woburn, MA, USA); ERBB2-neutralizing IgG (AF1129), ERBB3 (348-RB) and ERBB4 (1131-ER) Fc chimeric proteins were from R&D Systems.
In vivo spermatogenesis colony-forming assay
WT Sprague Dawley rats at 12 days of age were injected (i.p.) with 12 mg/kg busulfan (4 mg/ml in 50% DMSO) and then used as recipient males at 24 days of age. Busulfan is a spermatogonial toxin commonly used to kill spermatogonia in recipient rat testes prior to transplantation, because it increases the colonization efficiency by the donor stem cells.53,55,56 Donor cells were loaded into injection needles fashioned from 100 μl glass capillary tubes at 103 cells/65 μl SG Medium containing 0.032% (wt/vol) trypan blue, and then the entire volume was transplanted into the seminiferous tubules of anesthetized rats by retrograde injection through the rete testes.56,57 Recipient males were analyzed 28 days posttransplantation, and colonies of spermatogenesis expressing the tgGCS-EGFP transgene were scored using an IX70, Olympus fluorescence microscope (Olympus Inc.) equipped with the Simple-PCI software (C-Imaging Systems, Compix).54 Images of transgene expression in whole testes were captured using a Nikon SMZ1500 fluorescence stereomicroscope equipped with the ACT-1 imaging software (Nikon Instruments, Inc., Melville, NY, USA).54
Immunofluorescence labeling in testis sections
Testes were isolated from wild-type Sprague Dawley rats (Harlan, Inc.) and fixed for ~18 h at 4 °C in 0.1M sodium phosphate buffer, pH 7.2, containing 4% paraformaldehyde. Fixed testes were equilibrated through a 10, 18 and 25% sucrose (wt/v, dissolved in 1× PBS; Life Technologies, Inc., cat. no. 14040-182) gradient by sequential overnight incubations (~24 h) at 4 °C in 15 ml of each respective sucrose solution. Once equilibrated to 25% sucrose, testes were embedded in tissue-freezing medium (Electron Microscopy Sciences Inc., Hatfield, PA, USA, cat. no. 72592) and frozen using a Shandon Lips haw cryo-bath (Pittsburgh, PA, USA, cat. no. 45972). Frozen testes were used to prepare a parallel series of 8-μm cryo-sections. Frozen sections were stored at −40 °C until use in antibody-labeling assays. Prior to antibody labeling, sections were equilibrated in air to ~22–24 °C for 15 min, hydrated in PBS (Sigma, cat. no. D8537) at 22–24 °C for 10 min, heat treated at 80 °C for 8 min in 10 mM sodium citrate (pH 6.0) and then incubated for 1 h at 22–24 °C in blocking buffer (Roche Blocking Reagent (1% v/v) diluted in 0.1M sodium phosphate buffer, pH 7.2, containing Triton X100 (0.1% v/v)). Sections were then treated for 18–24 h at 22–24 °C with respective antibodies diluted in blocking buffer at the following concentrations: Anti-ERBB2, 0.25 μg/ml (Santa Cruz Inc., Dallas, TX, USA, Neu C18, sc-284); Anti-ERBB2, used at 1/150 dilution (Cell Signaling Technology Inc., Danvers, MA, USA, 29D8, 2165); Anti-PLZF, 0.2 μg/ml (EDM Millipore, Billerica, MA, USA, cat. no. OP128, clone ID 2A9); anti-SALL4, 0.2 μg/ml (Abnova cat. no. H0005716-M03, Clone ID 6E3); NRG1a1 (R&D Systems, Inc., cat. no. AF296NA); and NRG1b1 (R&D Systems, Inc., cat. no. AF396NA). After treatment with primary antibodies, sections were washed three times for 10 min/wash in 50 ml PBS and then incubated for 40 min at 22–24 °C with respective AlexaFluor594 (Life Technologies, Inc.), or AlexaFluor488 (Life Technologies, Inc.) secondary antibodies diluted to 4 μg/ml in PBS containing 5 μg/ml Hoechst 33342 dye (Molecular Probes, cat. no. H3570). After treatment with secondary antibodies, sections were washed three times at 10 min/wash in 50 ml PBS. After the third wash in PBS, sections were cover-slipped for viewing using Fluorogel mounting medium (Electron Microscopy Sciences, cat. no. 17985-10). Images were acquired using an IX70 Olympus fluorescence microscope (Olympus Inc.), as described above in 'Immunofluorescence labeling on seminiferous tubules' section. Labeled sections were cover-slipped using Fluorogel mounting medium and images were acquired as described above.
Immunofluorescence labeling in seminiferous tubule whole mounts
Seminiferous tubules were dissected from rat testes in 10-cm dishes containing Dulbecco modified Eagle’s medium: Ham’s F12 medium 1 : 1 (DHF12 Medium; Sigma, cat. no. D8437) at ~22–24 °C. Individual tubules were teased apart from testes by blunt dissection, rinsed in fresh DHF12 medium and fixed for 1 h at 4 °C in 20 ml 0.1M Sodium phosphate buffer, pH7.2, containing 4% paraformaldehyde. Optimal co-labeling with antibodies was obtained by suspending the paraformaldehyde-fixed tubules in ice-cold methanol and then immediately incubating them for additional 20 min at −20 °C (applied to obtain spermatogonial frequency data from 104–126d rats). Following fixation, tubules were washed three times in 20 ml PBS for ~10 min/wash at 22–24 °C and then stored at 4 °C for up to 1 week prior use. For indirect labeling with antibodies, 0.5–2.5 cm long tubule pieces were aliquoted into microfuge tubes containing 800 μl and 10 mM sodium citrate (pH 6.0) and incubated at 80 °C for 6–8 min. After heat treatment, tubules were immediately washed by dipping in PBS at 22–24 °C and then incubated for 1 h at 22–24 °C in blocking buffer before treating with respective primary and secondary antibodies as described above in 'Immunofluorescence labeling in testis' section.
EDU incorporation
Spermatogonia were passaged into SGF Medium and maintained for 48 h prior to treating cells with SGF Medium containing 3 μM ATRA for 4 days. EDU was added to culture medium at a final concentration of 10 μM and incubated for 7.5 h. Cells were fixed in 4% paraformaldehyde followed by methanol as described previously for immunocytochemistry. EDU was detected using the Click-it Edu Alexa-Fluor 594 Imaging Kit (Life Technologies Inc., cat. no. C10339).
Western blotting
Whole-cell lysates were prepared in lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630) with protease inhibitors (Complete EDTA-free protease inhibitor tablets, Roche Applied Science, Inc., Indianapolis, IN, USA, cat. no. 11836170001) and phosphatase inhibitors (PhosSTOP, Roche Applied Science, Inc., cat. no. 4906845001). Samples were clarified by centrifugation in a microfuge at 4 degrees for 10 min. Samples were run on SDS-PAGE gels under reducing conditions and transferred to nitrocellulose. Blots were probed for ERBB1 (Santa Cruz, Inc., cat. no. sc-03), ERBB2 (Lab Vision, Inc., cat. no. MS-599-P1), ERBB3 (Santa Cruz, Inc., cat. no. sc-285), ERBB4 (Cell Signaling Technology, Inc., cat. no. 4795), RET (R&D Systems, Inc., cat. no. AF482), NRG1α1 (R&D Systems, Inc., cat. no. AF296NA), NRG1β1 (R&D Systems, Inc., cat. no. AF396NA), phospho-AKT-S473 (Cell Signaling Technology, Inc., cat. no. 9271), phospho-P42/44 MAPK (Cell Signaling Technology, Inc., cat. no. 9101) and alpha-tubulin (Biogenex, Inc., Fremont, CA, USA, cat. no. MU121-UC). Secondary antibodies labeled with IRDye 800CW or 680LT were from Li-Cor Biosciences, Inc. (Lincoln, NB, USA). Blots were imaged on Odyssey Classic Quantitative Fluorescence Imaging System, Model 9120, also from Li-Cor Biosciences.
RT-PCR and qtPCR analysis of testis cell transcripts
Total RNA was isolated from testis cell sampless using the RNAqueous Micro Kit (Life Technologies, Inc.). RNA concentration in each isolated sample was measured using the RiboGreen Kit (Life Technologies, Inc.). Fifty nanograms of RNA from each cell sample was primed in a 20 μl reaction mix containing Oligo(dT) primers and Superscript III (Life Technologies, Inc.), according to the manufacturer’s protocol. Gene expression was analyzed by PCR using PfuTurbo in UltraHF Buffer from Stratagene, Inc. (Santa Clara, CA, USA), qtPCR conditions were as described,50 and RT-PCR conditions were as follows: 1 cycle, 2 min at 95°; 4 cycles 30 s at 95°, 45 s at 65°, 1 min at 68°; 32 cycles, 30 s at 95°, 45 s at 60°, and 1 min at 68°. PCR primers were designed to rat reference sequences in NCBI using Primer/Blast:
DR4 fibroblast feeder layer preparation
Primary stocks of DR4 MEFs were from ATCC, Inc. (Manassas, VA, USA), and expanded after plating into Dulbecco’s modified Eagle’s medium supplemented with 1.5 g/l sodium bicarbonate and 15% heat-inactivated FBS (MEF medium) at 37 °C/5% CO2 for up to four passages following their thawing and initial plating from the vial received from the manufacturer. Following expansion, secondary stocks of MEFs are irradiated (100 Gy) and then cryo-preserved in liquid nitrogen for future use in Recovery Cell Culture Freezing Medium (Life Technologies, Inc.) according to the manufacturer’s protocol. Tissue culture dishes were precoated with a solution of sterile 0.1% gelatin for 1 h at room temperature and rinsed once with sterile PBS before plating MEFs. Prior to use for culture with spermatogonia, irradiated MEFs were plated into gelatin-coated dishes (~6.8×104 cells/cm2) in MEF medium for 16–48 h, rinsed 1× with PBS and then preincubated in SG Medium for an additional 16–48 h. The SG Medium used for preincubation is then discarded and spermatogonia are passaged onto the MEFs in fresh SG Medium.
Abbreviations
- As:
-
A-single
- Apr:
-
A-paired
- Aal:
-
A-aligned
- AKT:
-
v-akt murine thymoma viral oncogene homolog 1
- AREG:
-
amphiregulin
- ATRA:
-
all-trans-retinoic acid
- BTC:
-
betacellulin
- CSPG5:
-
chondroitin sulfate proteoglycan 5
- DAZL:
-
Deleted in azoospermia
- EDTA:
-
ethylenediaminetetraacetic acid
- EDU:
-
5-ethynyl-2′-deoxyuridine
- EGF:
-
epidermal growth factor
- ERBB:
-
erythoblastoma virus B homolog
- FGF2:
-
fibroblast growth factor 2
- tgGCS-EGFP:
-
germ cell specific-enhanced green fluorescence protein-encoding transgene
- GDNF:
-
glial cell line derived neurotrophic factor
- GFRa1:
-
GDNF family receptor alpha 1
- HBE or HBEGF:
-
heparin-binding-epidermal growth factor
- IGEPAL:
-
octylphenoxypolyethoxyethanol
- KIT:
-
v-kit Hardy–Zuckerman 4 feline sarcoma viral oncogene homolog
- KITL:
-
KIT ligand
- MEF:
-
mouse embryonic fibroblast
- miRNA:
-
microRNA
- NRG:
-
neuregulin
- PBS:
-
phosphate-buffered saline
- qtPCR:
-
semi-quantitative polymerase chain reaction
- RT-PCR:
-
reverse transcription-polymerase chain reaction
- RET:
-
ret proto-oncogene
- SALL4:
-
spalt-like transcription factor 4
- SD medium:
-
spermatogonial differentiation medium
- SG medium:
-
spermatogonial culture medium
- TEX14:
-
testis expressed gene 14
- TGFα:
-
transforming growth factor alpha
- WNT:
-
wingless-type MMTV integration site family
- ZBTB16:
-
zinc finger and BTB domain containing 16.
References
Clermont Y . Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev 1972; 52: 198–236.
Braun RE, Behringer RR, Peschon JJ, Brinster RL, Palmiter RD . Genetically haploid spermatids are phenotypically diploid. Nature 1989; 337: 373–376.
Nagano M, Avarbock MR, Leonida EB, Brinster CJ, Brinster RL . Culture of mouse spermatogonial stem cells. Tissue Cell 1998; 30: 389–397.
Brinster RL, Zimmermann JW . Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA 1994; 91: 11298–11302.
Sato T, Katagiri K, Gohbara A, Inoue K, Ogonuki N, Ogura A et al. In vitro production of functional sperm in cultured neonatal mouse testes. Nature 2011; 471: 504–507.
Huckins C . The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec 1971; 169: 533–557.
Chan F, Oatley MJ, Kaucher AV, Yang QE, Bieberich CJ, Shashikant CS et al. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev 2014; 28: 1351–1362.
Nishimune Y, Haneji T, Kitamura Y . The effects of steel mutation on testicular germ cell differentiation. J Cell Physiol 1980; 105: 137–141.
Van Pelt AM, De Rooij DG . The origin of the synchronization of the seminiferous epithelium in vitamin A-deficient rats after vitamin A replacement. Biol Reprod 1990; 42: 677–682.
Monesi V . Autoradiographic study of DNA synthesis and the cell cycle in spermatogonia and spermatocytes of mouse testis using tritiated thymidine. J Cell Biol 1962; 14: 1–18.
Dym M, Fawcett DW . Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod 1971; 4: 195–215.
Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287: 1489–1493.
Takashima S, Kanatsu-Shinohara M, Tanaka T, Morimoto H, Inoue K, Ogonuki N et al. Functional Differences between GDNF-Dependent and FGF2-Dependent Mouse Spermatogonial Stem Cell Self-Renewal. Stem cell reports 2015; 4: 489–502.
Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 2003; 69: 612–616.
Kubota H, Avarbock MR, Brinster RL . Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci USA 2004; 101: 16489–16494.
Ryu BY, Kubota H, Avarbock MR, Brinster RL . Conservation of spermatogonial stem cell self-renewal signaling between mouse and rat. Proc Natl Acad Sci USA 2005; 102: 14302–14307.
Hamra FK, Chapman KM, Nguyen DM, Williams-Stephens AA, Hammer RE, Garbers DL . Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc Natl Acad Sci USA 2005; 102: 17430–17435.
Kanatsu-Shinohara M, Muneto T, Lee J, Takenaka M, Chuma S, Nakatsuji N et al. Long-term culture of male germline stem cells from hamster testes. Biol Reprod 2008; 78: 611–617.
Kanatsu-Shinohara M, Miki H, Inoue K, Ogonuki N, Toyokuni S, Ogura A et al. Long-term culture of mouse male germline stem cells under serum-or feeder-free conditions. Biol Reprod 2005; 72: 985–991.
Yeh JR, Zhang X, Nagano MC . Indirect effects of Wnt3a/beta-catenin signalling support mouse spermatogonial stem cells in vitro. PLoS One 2012; 7: e40002.
Yeh JR, Zhang X, Nagano MC . Wnt5a is a cell-extrinsic factor that supports self-renewal of mouse spermatogonial stem cells. J Cell Sci 2011; 124 (Pt 14 2357–2366.
Kissel H, Timokhina I, Hardy MP, Rothschild G, Tajima Y, Soares V et al. Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. EMBO J 2000; 19: 1312–1326.
Geissler EN, McFarland EC, Russell ES . Analysis of pleiotropism at the dominant white-spotting (W) locus of the house mouse: a description of ten new W alleles. Genetics 1981; 97: 337–361.
Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM . Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 1999; 140: 5894–5900.
Manova K, Nocka K, Besmer P, Bachvarova RF . Gonadal expression of c-kit encoded at the W locus of the mouse. Development 1990; 110: 1057–1069.
Morales C, Griswold MD . Retinol-induced stage synchronization in seminiferous tubules of the rat. Endocrinology 1987; 121: 432–434.
Haneji T, Maekawa M, Nishimune Y . Retinoids induce differentiation of type A spermatogonia in vitro: organ culture of mouse cryptorchid testes. J Nutr 1983; 113: 1119–1123.
Steinberger E, Steinberger A, Perloff WH . Initiation of spermatogenesis in vitro. Endocrinology 1964; 74: 788–792.
Kierszenbaum AL . Mammalian spermatogenesis in vivo and in vitro: a partnership of spermatogenic and somatic cell lineages. Endocr Rev 1994; 15: 116–134.
Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM et al. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod 2008; 78: 537–545.
Dann CT, Alvarado AL, Molyneux LA, Denard BS, Garbers DL, Porteus MH . Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells 2008; 26: 2928–2937.
Chapman KM, Medrano GA, Jaichander P, Chaudhary J, Waits AE, Nobrega MA et al. Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Rep 2015; 10: 1828–1835.
Zhang J, Eto K, Honmyou A, Nakao K, Kiyonari H, Abe S . Neuregulins are essential for spermatogonial proliferation and meiotic initiation in neonatal mouse testis. Development 2011; 138: 3159–3168.
Galvez-Contreras AY, Quinones-Hinojosa A, Gonzalez-Perez O . The role of EGFR and ErbB family related proteins in the oligodendrocyte specification in germinal niches of the adult mammalian brain. Front Cell Neurosci 2013; 7: 258.
Abid SN, Richardson TE, Powell HM, Jaichander P, Chaudhary J, Chapman KM et al. A-single spermatogonia heterogeneity and cell cycles synchronize with rat seminiferous epithelium stages VIII-IX. Biol Reprod 2014; 90: 32.
Tokita Y, Keino H, Matsui F, Aono S, Ishiguro H, Higashiyama S et al. Regulation of neuregulin expression in the injured rat brain and cultured astrocytes. J Neurosci 2001; 21: 1257–1264.
Lemmens K, Doggen K, De Keulenaer GW . Activation of the neuregulin/ErbB system during physiological ventricular remodeling in pregnancy. Am J Physiol Heart Circ Physiol 2011; 300: H931–H942.
Shuo T, Aono S, Nakanishi K, Tokita Y, Kuroda Y, Ida M et al. Ectodomain shedding of neuroglycan C, a brain-specific chondroitin sulfate proteoglycan, by TIMP-2- and TIMP-3-sensitive proteolysis. J Neurochem 2007; 102: 1561–1568.
Cronkhite JT, Norlander C, Furth JK, Levan G, Garbers DL, Hammer RE . Male and female germline specific expression of an EGFP reporter gene in a unique strain of transgenic rats. Dev Biol 2005; 284: 171–183.
Vincent S, Segretain D, Nishikawa S, Nishikawa SI, Sage J, Cuzin F et al. Stage-specific expression of the Kit receptor and its ligand (KL) during male gametogenesis in the mouse: a Kit-KL interaction critical for meiosis. Development 1998; 125: 4585–4593.
Packer AI, Besmer P, Bachvarova RF . Kit ligand mediates survival of type A spermatogonia and dividing spermatocytes in postnatal mouse testes. Mol Reprod Dev 1995; 42: 303–310.
Yoshinaga K, Nishikawa S, Ogawa M, Hayashi S, Kunisada T, Fujimoto T et al. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development 1991; 113: 689–699.
Yan W, Suominen J, Toppari J . Stem cell factor protects germ cells from apoptosis in vitro. J Cell Sci 2000; 113: 161–168.
Kubota H, Avarbock MR, Schmidt JA, Brinster RL . Spermatogonial stem cells derived from infertile Wv/Wv mice self-renew in vitro and generate progeny following transplantation. Biol Reprod 2009; 81: 293–301.
Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA et al. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev Biol 2015; 397: 140–149.
yang QE, Racicot KE, Kaucher AV, Oatley MJ, Oatley JM . MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development 2013; 140: 280–290.
Huszar JM, Payne CJ . MicroRNA 146 (Mir146) modulates spermatogonial differentiation by retinoic acid in mice. Biol Reprod 2013; 88: 15.
Hamra FK, Chapman KM, Nguyen D, Garbers DL . Identification of neuregulin as a factor required for formation of aligned spermatogonia. J Biol Chem 2007; 282: 721–730.
Chapman KM, Saidley-Alsaadi D, Syvyk AE, Shirley JR, Thompson LM, Hamra FK. Spermatogonial stem cell mediated gene transfer. In: Pease S, Saunders T (eds). Advanced Protocols for Animal Transgenesis. Springer-Verlag: Berlin, Heidelberg, 2011, pp 237–66.
Wu Z, Falciatori I, Molyneux LA, Richardson TE, Chapman KM, Hamra FK . Spermatogonial culture medium: an effective and efficient nutrient mixture for culturing rat spermatogonial stem cells. Biol Reprod 2009; 81: 77–86.
Clermont Y, Perey B . Quantitative study of the cell population of the seminiferous tubules in immature rats. Am J Anat 1957; 100: 241–267.
Hamra FK, Chapman KM, Wu Z, Garbers DL . Isolating highly pure rat spermatogonial stem cells in culture. Methods Mol Biol 2008; 450: 163–179.
Hamra FK, Gatlin J, Chapman KM, Grellhesl DM, Garcia JV, Hammer RE et al. Production of transgenic rats by lentiviral transduction of male germ-line stem cells. Proc Natl Acad Sci USA 2002; 99: 14931–14936.
Hamra FK, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE et al. Defining the spermatogonial stem cell. Dev Biol 2004; 269: 393–410.
Ryu BY, Orwig KE, Avarbock MR, Brinster RL . Stem cell and niche development in the postnatal rat testis. Dev Biol 2003; 263: 253–263.
Ogawa T, Dobrinski I, Brinster RL . Recipient preparation is critical for spermatogonial transplantation in the rat. Tissue Cell 1999; 31: 461–472.
Dym M . The mammalian rete testis--a morphological examination. Anat Rec 1976; 186: 493–523.
Pankonin MS, Gallagher JT, Loeb JA . Specific structural features of heparan sulfate proteoglycans potentiate neuregulin-1 signaling. J Biol Chem 2005; 280: 383–388.
Zhang Z, Prentiss L, Heitzman D, Stahl RC, DiPino F Jr, Carey DJ . Neuregulin isoforms in dorsal root ganglion neurons: effects of the cytoplasmic domain on localization and membrane shedding of Nrg-1 type I. J Neurosci Res 2006; 84: 1–12.
Acknowledgements
We thank Priscilla Jaichander and Tuytanh Nguyen for their help with this study. This work was supported by National Institutes of Health grants from The Eunice Kennedy Shriver National Institute of Child Health and Human Development: R01HD053889 and R01HD061575, and The Office of the Director: R24OD011108.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chapman, K., Medrano, G., Chaudhary, J. et al. NRG1 and KITL signal downstream of retinoic acid in the germline to support soma-free syncytial growth of differentiating spermatogonia. Cell Death Discovery 1, 15018 (2015). https://doi.org/10.1038/cddiscovery.2015.18
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/cddiscovery.2015.18
This article is cited by
-
Expression profile of microRNAs in the testes of patients with Klinefelter syndrome
Scientific Reports (2020)
-
Scaffold protein Lin7 family in membrane skeletal protein complex in mouse seminiferous tubules
Histochemistry and Cell Biology (2019)
-
Discovering in vitro spermatogenesis stimulating factors
Cell Death & Disease (2015)