Abstract
The chicken anaemia virus-derived protein Apoptin/VP3 (CAV-Apoptin) has the important ability to induce tumour-selective apoptosis in a variety of human cancer cells. Recently the first human Gyrovirus (HGyV) was isolated from a human skin swab. It shows significant structural and organisational resemblance to CAV and encodes a homologue of CAV-Apoptin/VP3. Using overlapping primers we constructed a synthetic human Gyrovirus Apoptin (HGyV-Apoptin) fused to green fluorescent protein in order to compare its apoptotic function in various human cancer cell lines to CAV-Apoptin. HGyV-Apoptin displayed a similar subcellular expression pattern as observed for CAV-Apoptin, marked by translocation to the nucleus of cancer cells, although it is predominantly located in the cytosol of normal human cells. Furthermore, expression of either HGyV-Apoptin or CAV-Apoptin in several cancer cell lines triggered apoptosis at comparable levels. These findings indicate a potential anti-cancer role for HGyV-Apoptin.
Similar content being viewed by others
Main
Apoptin was originally identified as the death-inducing protein VP3 from chicken anaemia virus (CAV), the first member of the Gyrovirus genus. Infection with the virus, which is mainly transmitted through contaminated feathers, has been shown to trigger apoptosis in thymocytes and myeloid progenitor cells of young chicken.1 CAV is a single-stranded DNA virus which encodes three proteins, VP1 (capsid protein), VP2 (protein phosphatase and scaffold protein) and VP3.2 In vitro, expression of VP3 alone induces apoptosis of chicken lymphoblastoid T-cells and myeloid cells in a similar way as observed after CAV infection.3, 4 Hence, VP3 was renamed Apoptin.
CAV-Apoptin is a 121 amino-acid protein rich in proline, serine, threonine and basic amino acids.5, 6 It contains a bipartite-type nuclear localisation signal (NLS1 and NLS2) at position 82-88 and 111-121 and a nuclear export signal (NES) suggesting potential shuttling of the protein between nucleus and cytoplasm.7, 8 Both predicted NLSs of Apoptin are required for its efficient nuclear localisation.7 Furthermore, Apoptin contains several potential phosphorylation sites, including threonine 108.9
Importantly, Apoptin has the ability to induce apoptosis in various human cancer cells but not in normal cells.10, 11, 12 It triggers apoptosis through the intrinsic mitochondrial pathway requiring caspase-3 and caspase-9, but not caspase-8. Cell death is induced independently of p53 but possibly involves the p53 family member p73 that can transactivate pro-apoptotic targets such as Bax and Puma.13, 14 Furthermore, the sphingomyelin-ceramide pathway has been shown to have a role in Apoptin-induced cytotoxicity.15 However, the precise cellular mechanisms of Apoptin-induced cell death and the mode of tumour selectivity remain unclear. There are distinct differences regarding Apoptin expression between normal and tumour cells, which may provide clues about the mechanisms of tumour cell sensitivity to Apoptin. Firstly, in tumour cells Apoptin localises to the nucleus, which is important for its ability to induce apoptosis, whereas in normal cells it is predominantly expressed in the cytoplasm.10, 16 In addition, Apoptin is phosphorylated on threonine 108 in tumour but not in normal cells, which drives nuclear accumulation in tumour cells.9 The specific kinase/s responsible for Apoptin phosphorylation remain unidentified. However, several kinases, including PKCβ and CDK2 have been shown to phosphorylate Apoptin.17, 18
Recently, a virus showing significant homology to CAV was isolated from skin swabs of healthy volunteers and designated human Gyrovirus or HGyV.19 Similar to CAV the HGyV genome consists of a single-stranded negative-sense circular DNA of 2.315 nucleotides (compared with 2.290–2.320 for CAV), containing three partially overlapping open reading frames. Alignment with CAV revealed relatively low overall sequence identity with a maximal identity of 70% in the region of nucleotides 100–700 but a similar organisation of the promoter region and the encoded proteins. In addition to VP1 and VP2 HGyV encodes a slightly larger, 125 amino-acid homologue of CAV-Apoptin/VP3 (121 amino acids for comparison). Despite a low overall identity important regions such as the nuclear localisation and export signals and phosphorylation sites are conserved between HGyV- and CAV-Apoptin. The effects or involvement of this novel virus in human disease and the functional properties of human Gyrovirus Apoptin (HGyV-Apoptin) are currently unknown.
To investigate whether HGyV-Apoptin has apoptotic and tumour selective activity similar to its homologue CAV-Apoptin we developed a synthetic HGyV-Apoptin fused to green fluorescent protein (GFP). Expression of this construct in human cancer cell lines revealed a subcellular localisation and pro-apoptotic function comparable to CAV-Apoptin.
Results
Expression of HGyV-Apoptin in human cancer cells
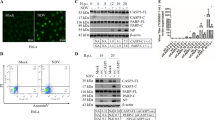
To test the expression of the newly generated HGyV-Apoptin constructs, HCT116 colon carcinoma and Saos-2 osteosarcoma cells were transfected with the corresponding plasmids pHGyV-GFP-AP and pHGyV-FLAG-AP. Western blot analysis of transfected cells after 2 days revealed expression of both HGyV-GFP-AP and HGyV-FLAG-AP that could be detected at the estimated molecular weights (Figures 1a and b). Compared with its CAV homologue HGyV-GFP-AP is slightly larger as expected from the difference in length. Re-probing with a CAV-Apoptin phospho-specific antibody directed against phosphorylated threonine 108 did not reveal any cross-reaction with HGyV-Apoptin (data not shown).
Expression of CAV- and HGyV-Apoptin in cancer cells. (a) Alignment of the protein sequence of CAV-Apoptin (NP_056774.1) and HGyV-Apoptin (CBZ41794.1) using computer software Align (http://xylian.igh.cnrs.fr/bin/align-guess.cgi). Important functional domains, including LRS (leucine-rich domain), NLS1/2 and NES are indicated by boxes and the predicted phosphorylation sites threonine 108 or threonine 111 are indicated by black arrows. Whole cell lysates of HCT116 colon carcinoma (b) and Saos-2 osteosarcoma (c) cells transfected with the indicated plasmids were prepared and western blot analysis for detection of GFP- or FLAG-Apoptin, respectively, as well as β-Actin as a loading control was performed. Molecular weights of the bands are indicated with arrows and blots were cut and combined at the black line
HGyV-Apoptin translocates to the nucleus and induces changes in nuclear morphology in cancer cells
CAV-Apoptin is known to translocate to the nucleus in transformed cells which is at least partially important for the induction of apoptosis in these cells. To investigate the subcellular distribution of HGyV-Apoptin, which also contains putative nuclear localisation and export signals (Figure 1a), as well as morphological changes caused by its expression, cells were transfected with different Apoptin constructs and analysed by fluorescence microscopy. Imaging of HCT116 colon carcinoma cells transfected with pCAV-GFP-AP or pHGyV-GFP-AP at 24 and 48 h post transfection revealed a similar expression pattern of CAV- and HGyV-Apoptin (Figure 2a). CAV-Apoptin has been shown to initially exhibit a fine granular distribution within the nucleus, later clustering to form aggregates, at which point the cell becomes apoptotic. This characteristic morphology was also observed with HGyV-Apoptin in transfected Saos-2 osteosarcoma cells after 2 and 5 days, respectively (Figure 2b). To exclude a potential promoting effect of the GFP-tag on nuclear translocation Saos-2 cells were transfected with FLAG-CAV-AP or FLAG-HGyV-AP. Subsequent indirect immunofluorescence to detect FLAG-Apoptin revealed the same expression pattern as observed for GFP-Apoptin (Figure 2c). Similar cellular distribution was observed in the HT1080 breast cancer cell line (data not shown).
Nuclear translocation of GFP- and FLAG-Apoptin in cancer cells. HCT116 (a) and Saos-2 (b) cancer cells were transfected with pCAV-GFP-AP or pHGyV-GFP-AP and the corresponding pEGFP-C1 control plasmid. Cells were directly imaged in culture dishes after 1 or 2 days (a) or fixed and counterstained with DAPI at 2 and 5 days post-transfection (b) as indicated. Saos-2 cells (c) transfected with pCAV-FLAG-AP or pHGyV-FLAG-AP were fixed after 5 days and stained with a primary mouse anti-FLAG and secondary FITC anti-mouse antibody for detection of FLAG-Apoptin. Nuclei were detected by counterstaining with DAPI. Arrows indicate apoptotic nuclei
HGyV-GFP-Apoptin induces cell death in Saos-2 osteosarcoma cells
To examine the cytotoxic function of HGyV-GFP-AP Saos-2 human osteosarcoma cells were transfected by nucleofection with pHGyV-GFP-AP, pCAV-GFP-AP or the corresponding pEGFP-C1 control vector. Apoptosis was quantified by flow cytometric analysis as percentage of Annexin-V-positive cells among the transfected cells after Annexin-V-APC/ propidium iodide (PI) staining. After 5 days between 40 and 50% of cells expressing GFP-Apoptin were apoptotic compared with only 5% apoptotic cells in those transfected with pEGFP-C1 (Figure 3a). The level of cell death induced by HGyV-Apoptin was similar to the level achieved by CAV-Apoptin.
Induction of cell death in Saos-2 cells at 5 days after transfection. Saos-2 cells were transfected by nucleofection with pCAV-GFP-AP, pHGyV-GFP-AP or the corresponding pEGFP-C1 control plasmid, as well as pCAV-FLAG-AP and pHGyV-FLAG-AP. Cell death was measured by flow cytometric analysis after Annexin-V-APC/ PI staining. (a) Quantification of cell death as % of APC-labelled cells in the GFP-positive population after 5 days (*P<0.01). Error bars indicate standard deviation of three experiments. (b) Corresponding FACS dot plots of Annexin-V-APC (x-axis) versus PI (y-axis) obtained after 2 and 5 days. (c) After 5 days transfected Saos-2 cells were fixed, stained with a primary mouse anti-FLAG and secondary FITC anti-mouse antibody (for pCAV-FLAG-AP and pHGyV-FLAG-AP) and counterstained with DAPI for the detection of nuclear morphology. Cell death was quantified as the percentage of GFP- or FLAG-positive cells showing condensed or fragmented nuclei. Error bars indicate standard deviation of two independent experiments
Furthermore, a colony-forming assay of HT1080 cells comparably transfected with pEGFP-C1 or pHGyV-GFP-AP showed very few HGyV-GFP-AP expressing cells, whereas the control pEGFP-C1 transfected cells were able to form countless colonies in the presence of G418 (data not shown) indicating a cytotoxic effect of HGyV-GFP-AP.
To compare the pro-apoptotic effect of GFP- and FLAG-tagged Apoptin, Saos-2 cells were transfected with different Apoptin and control plasmids. After 5 days cells were fixed, stained with anti-FLAG antibody (for pCAV-FLAG-AP and pHGyV-FLAG-AP) and counterstained with DAPI for the detection of nuclear morphology. Apoptosis was quantified by scoring cells expressing either GFP- or FLAG-Apoptin and displaying clear apoptotic nuclear morphology. Results suggested that both CAV- and HGyV-Apoptin efficiently induce apoptosis as compared with the pEGFP-C1 control. Furthermore, no significant differences between the GFP- or FLAG-tagged Apoptin could be observed (Figure 3c).
HGyV-GFP-Apoptin is expressed in the cytoplasm of normal human fibroblasts and translocates to the nucleus of transformed counterparts
One of the important characteristics of CAV-Apoptin is its distinct tumour specific nuclear localisation, which is necessary for its ability to selectively induce apoptosis in transformed but not in normal cells. To investigate whether HGyV-Apoptin shows the same tumour selectivity pEGFP-C1, pHGyV-GFP-AP, pCAV-GFP-AP, pHGyV-FLAG-AP and pCAV-FLAG-AP were transfected into 1BR3 normal human fibroblasts and their SV40-LT transformed counterparts, (1BR3LT). After 3 days cells were counterstained with DAPI and analysed by fluorescent microscopy to quantify apoptosis by scoring the GFP-positive cells containing condensed or fragmented nuclei. The normal 1BR3 fibroblasts exhibited a marked difference in cellular localisation of both CAV-GFP-AP and HGyV-GFP-AP as compared with transformed 1BR3LT (Figure 4a). In normal cells, HGyV- and CAV-GFP-Apoptin were mainly expressed in the cytoplasm, while in the transformed cells both CAV- and HGyV-GFP-Apoptin had a predominantly nuclear localisation. Similarly, FLAG-tagged CAV- or HGyV-Apoptin was localised to the nucleus of transformed 1BR3LT cells, whereas it was predominantly cytoplasmic in normal 1BR3 fibroblasts (data not shown). This difference in subcellular localisation was associated with increased apoptosis in the transformed cells transduced with either FLAG- or GFP-Apoptin (Figure 4b).
Cytosolic expression of GFP-Apoptin and quantification of apoptosis in 1BR3 normal human fibroblasts. Normal (1BR3) and transformed (1BR3LT) human fibroblast cell lines were transfected by nucleofection with pCAV-GFP-AP, pHGyV-GFP-AP or the corresponding pEGFP-C1 control plasmid as well as pCAV-FLAG-AP and pHGyV-FLAG-AP. After 3 days cells were fixed and counterstained with DAPI to analyse the subcellular localisation of GFP-Apoptin. Expression of FLAG-AP was detected using primary mouse anti-FLAG and secondary FITC anti-mouse antibody (a). Apoptosis was quantified by scoring GFP- or FLAG-positive cells containing condensed or fragmented nuclei (b). Over one hundred transfected cells were analysed and experiments were repeated twice (error bars indicate standard deviation)
Discussion
Tumour specificity remains one of the main hurdles for successful treatment of cancer.20 A number of viral and cellular proteins have been described that show tumour-specific toxicity, including Apoptin, Trail, mda-7 (etc).21 The therapeutic potential of such proteins and their cellular targets is of immense interest. Recently, a HGyV has been identified encoding a viral protein resembling the CAV-derived Apoptin. Here we provide clear evidence that this HGyV-Apoptin shows a tumour-specific nuclear expression pattern as has been described previously for CAV-Apoptin to be important for its cytotoxicity.7 Furthermore, expression of HGyV-Apoptin resulted in the induction of apoptosis in transformed cells but not in the normal human cell line 1BR3. However, further studies, including the use of different types of normal cells, are necessary to confirm that HGyV-Apoptin has tumour-selective cytotoxicity and could thus provide a novel tool for cancer treatment.
Many studies in the past decade have provided insight into the mechanism of Apoptin-induced apoptosis. However, the basis for its increased sensitivity for cancer cells remains largely unclear. In addition to translocation to the nucleus, CAV-Apoptin is phosphorylated on threonine 108 in cancer cells but not in normal cells, which seems to be important for enabling Apoptin to induce apoptosis.9 Several cellular kinases have been proposed to be responsible for CAV-Apoptin phosphorylation.14, 18 We have recently identified PKCβ to be important for phosphorylation and activation of CAV-Apoptin.17 It would be important to investigate whether HGyV-Apoptin is phosphorylated in a similar manner by the kinases which have been shown to interact with CAV-Apoptin. These studies are currently on-going.
Although so far no link between this novel HGyV and any human disease has been identified, the clear apoptotic activity of its encoded protein HGyV-Apoptin offers an intriguing opportunity to investigate its potential pathological role. Furthermore a better understanding of the function of the viral protein HGyV-Apoptin may reveal involvement in human disease and lead to the development of novel therapeutics for cancer.
Materials and Methods
Cell lines and cell culture
HCT116 human colorectal carcinoma cells were obtained from ATCC (Manassas, VA, USA) and maintained in McCoy’s 5A medium supplemented with 10% foetal bovine serum 40 U/ml Penicillin and 40 μg/ml Streptomycin (all PAA Laboratories, Yeovil, UK). Saos-2 human osteosarcoma cells were purchased from ATCC and grown in DMEM (PAA Laboratories) supplemented with 10% foetal bovine serum 40 U/ml Penicillin and 40 μg/ml Streptomycin. 1BR3 normal and 1BR3 SV40 1BR3LT were a gift from Prof. Alan Lehmann (Sussex Centre for Genome Damage and Stability, University of Sussex, UK) and maintained in DMEM containing 15% foetal bovine serum 40 U/ml Penicillin and 40 μg/ml Streptomycin. All cell lines were incubated at 37 °C and 5% CO2.
Generation of HGyV-Apoptin
A codon optimised HGyV-Apoptin gene was constructed using primers overlapping by 15 bases (Table 1). FOR1 introduces a Kozak sequence upstream of the ATG start codon and a BamHI site for subsequent cloning. REV5 introduces an EcoRI site for subsequent cloning. Primers FOR1-5 and REV1-4 were mixed in equimolar amounts and heated to 95 0C for 2 min and cooled slowly to room temperature. Gaps were filled in with T4 DNA Polymerase for 15 min at 12 °C and finally the gaps were closed by treatment with T4 DNA Ligase for 15 min at RT. The resulting product was PCR amplified with primers FOR1 and REV5 and the PCR product was TOPO TA cloned (Invitrogen Life Technologies, Paisley, UK).
Inserts were sequence verified and a correct HGyV-AP was removed from the TOPO vector by BamHI/EcoRI digestion and cloned in-frame with GFP into the BglII/EcoRI digested mammalian expression vector pEGFP-C1 (Clontech, Saint-Germain-en-Laye, France) and named pHGyV-GFP-AP.
pHGyV-FLAG-AP was cloned by removing HGyV-AP from the TOPO vector by EcoRI digestion and filling in with Klenow. p3X-FLAG (Sigma-Aldrich, Poole, UK) was digested with BamHI and EcoRI, blunt ended with Mungbean nuclease and phosphates were removed by calf intestinal phosphatase treatment. Vector and insert were blunt end ligated and the correct orientation was determined by BstXI and BamHI digestion. Plasmids containing an insert in the correct orientation were sequence verified.
Plasmids and transient transfection
Expression vectors for HGyV-Apoptin (pHGyV-GFP-AP, pHGyV-FLAG-AP) were constructed as part of this study. pCAV-GFP-AP was generated as described previously10 and pCAV-FLAG-AP was received from Dr. Jose Teodoro (Goodman Cancer Research Centre, Montreal, QC, Canada). Other control vectors used were pcDNA3.1 (Invitrogen) and pEGFP-C1.
HCT116 cells were transiently transfected using X-tremeGENE HP transfection reagent according to the manufacturer’s protocol (Roche Diagnostics, Burgess Hill, UK). Transfection of Saos-2 and 1BR3/1BR3LT cells was achieved using Nucleofector technology as recommended by the manufacturer (Lonza, Basel, Switzerland).
Flow cytometric analysis
Cells were seeded on 12-well plates and collected at 2 or 5 days after nucleofection. Apoptosis was quantified by staining with Annexin-V-APC (Invitrogen) and PI (Sigma) and subsequent analysis on a BD FACSCanto II (Becton Dickinson, Oxford, UK). Data was analysed using FlowJo software (Tree Star, Inc., Ashland, OR, USA).
Western blot analysis
Protein extracts were prepared by washing cells and directly lysing them in Laemmli sample buffer (62.5 mM Tris-HCl pH 6.7, 100 mm β-mercaptoethanol, 2% SDS, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 100 μg/ml phenylmethylsulphoxide). SDS-PAGE was performed and proteins were transferred to nitrocellulose membranes (Sigma). Membranes were incubated with specific primary and secondary antibodies as indicated and developed using a home-made enhanced chemiluminescent system. The antibodies used were: rabbit anti-GFP (Cell Signaling, Beverly, MA, USA), mouse anti-FLAG (Sigma), mouse anti-β-Actin (Sigma) and secondary anti-mouse (Sigma) or anti-rabbit (GE Healthcare, Chalfont St. Giles, UK) linked to horseradish peroxidase.
Fluorescence microscopy
HCT116 cells were imaged directly on culture plates at 1 or 2 days post-transfection using an Olympus CKX41 inverted microscope (Southend-on-Sea, UK).
Saos-2 and 1BR3/1BR3LT cells were transfected by nucleofection and grown on 8-well culture slides (Becton Dickinson). After 3 or 5 days cells were washed once with PBS, fixed in 2% paraformaldehyde for 30 min at 37 °C and permeabilized with 0.2% Triton X-100 for 20 min. For the detection of FLAG tag cells were washed in PBS, blocked using 3% BSA in PBS-T for 30 min at 37 °C and incubated with the primary anti-FLAG antibody and the secondary antibody anti-mouse-FITC (both Sigma) for 2 h or 1 h, respectively, at 37 °C with washing steps in PBS between the incubations. All cells were mounted in Vectashield mounting medium containing DAPI (Vector Laboratories, Peterborough, UK) for nuclear staining and images were acquired using an Olympus BX61 automated fluorescence microscope.
Apoptosis was quantified by scoring the GFP-positive cells containing condensed or fragmented nuclei. Over one hundred transfected cells were analysed and experiments were repeated twice.
Statistical analysis
A paired two-tailed Student’s t-test was used to calculate significance.
Abbreviations
- CAV:
-
chicken anaemia virus
- HGyV:
-
human Gyrovirus
- GFP:
-
green fluorescent protein
- NLS:
-
nuclear localisation signal
- NES:
-
nuclear export signal
- LRS:
-
leucine rich stretch
- PKCβ:
-
protein kinase c beta
- CDK2:
-
cyclin dependent kinase 2
- HGyV-AP:
-
HGyV-derived Apoptin
- GFP-AP:
-
GFP-tagged Apoptin
- FLAG-AP:
-
FLAG-tagged Apoptin
- PI:
-
propidium iodide
- APC:
-
allophycocyanin
- 1BR3LT:
-
SV40 large T transformed 1BR3 fibroblasts
- FITC:
-
fluorescein isothiocyanate
- PBS:
-
phosphate-buffered saline
- SDS:
-
sodium dodecyl sulphate
- BSA:
-
bovine serum albumin
- ECL:
-
enhanced chemiluminescence
- CIP:
-
calf intestinal phosphatase
References
Noteborn MH, de Boer GF, van Roozelaar DJ, Karreman C, Kranenburg O, Vos JG et al. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol 1991; 65: 3131–3139.
Noteborn MH, Koch G . Chicken anaemia virus infection: molecular basis of pathogenicity. Avian Pathol 1995; 24: 11–31.
Noteborn MH, Todd D, Verschueren CA, de Gauw HW, Curran WL, Veldkamp S et al. A single chicken anemia virus protein induces apoptosis. J Virol 1994; 68: 346–351.
Tavassoli M, Guelen L, Luxon BA, Gaken J . Apoptin: specific killer of tumor cells? Apoptosis 2005; 10: 717–724.
Backendorf C, Visser AE, de Boer AG, Zimmerman R, Visser M, Voskamp P et al. Apoptin: therapeutic potential of an early sensor of carcinogenic transformation. Annu Rev Pharmacol Toxicol 2008; 48: 143–169.
Pietersen AM, van der Eb MM, Rademaker HJ, van den Wollenberg DJ, Rabelink MJ, Kuppen PJ et al. Specific tumor-cell killing with adenovirus vectors containing the apoptin gene. Gene Ther 1999; 6: 882–892.
Danen-Van Oorschot AA, Zhang YH, Leliveld SR, Rohn JL, Seelen MC, Bolk MW et al. Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J Biol Chem 2003; 278: 27729–27736.
Poon IK, Oro C, Dias MM, Zhang J, Jans DA . Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res 2005; 65: 7059–7064.
Rohn JL, Zhang YH, Aalbers RI, Otto N, Den Hertog J, Henriquez NV et al. A tumor-specific kinase activity regulates the viral death protein Apoptin. J Biol Chem 2002; 277: 50820–50827.
Guelen L, Paterson H, Gaken J, Meyers M, Farzaneh F, Tavassoli M . TAT-apoptin is efficiently delivered and induces apoptosis in cancer cells. Oncogene 2004; 23: 1153–1165.
Zhang YH, Kooistra K, Pietersen A, Rohn JL, Noteborn MH . Activation of the tumor-specific death effector apoptin and its kinase by an N-terminal determinant of simian virus 40 large T antigen. J Virol 2004; 78: 9965–9976.
Zhuang SM, Shvarts A, van Ormondt H, Jochemsen AG, van der Eb AJ, Noteborn MH . Apoptin, a protein derived from chicken anemia virus, induces p53-independent apoptosis in human osteosarcoma cells. Cancer Res 1995; 55: 486–489.
Klanrit P, Flinterman MB, Odell EW, Melino G, Killick R, Norris JS et al. Specific isoforms of p73 control the induction of cell death induced by the viral proteins, E1A or apoptin. Cell Cycle 2008; 7: 205–215.
Maddika S, Booy EP, Johar D, Gibson SB, Ghavami S, Los M . Cancer-specific toxicity of apoptin is independent of death receptors but involves the loss of mitochondrial membrane potential and the release of mitochondrial cell-death mediators by a Nur77-dependent pathway. J Cell Sci 2005; 118: 4485–4493.
Liu X, Zeidan YH, Elojeimy S, Holman DH, El-Zawahry AM, Guo GW et al. Involvement of sphingolipids in apoptin-induced cell killing. Mol Ther 2006; 14: 627–636.
Zhang YH, Leliveld SR, Kooistra K, Molenaar C, Rohn JL, Tanke HJ et al. Recombinant Apoptin multimers kill tumor cells but are nontoxic and epitope-shielded in a normal-cell-specific fashion. Exp Cell Res 2003; 289: 36–46.
Jiang J, Cole D, Westwood N, Macpherson L, Farzaneh F, Mufti G et al. Crucial roles for protein kinase C isoforms in tumor-specific killing by apoptin. Cancer Res 2010; 70: 7242–7252.
Maddika S, Panigrahi S, Wiechec E, Wesselborg S, Fischer U, Schulze-Osthoff K et al. Unscheduled Akt-triggered activation of cyclin-dependent kinase 2 as a key effector mechanism of apoptin's anticancer toxicity. Mol Cell Biol 2009; 29: 1235–1248.
Sauvage V, Cheval J, Foulongne V, Gouilh MA, Pariente K, Manuguerra JC et al. Identification of the first human gyrovirus, a virus related to chicken anemia virus. J Virol 2011; 85: 7948–7950.
Flinterman M, Farzaneh F, Habib N, Malik F, Gaken J, Tavassoli M . Delivery of therapeutic proteins as secretable TAT fusion products. Mol Ther 2009; 17: 334–342.
Argiris K, Panethymitaki C, Tavassoli M . Naturally occurring, tumor-specific, therapeutic proteins. Exp Biol Med (Maywood) 2011; 236: 524–536.
Acknowledgements
DC is supported by a Cancer Research UK Studentship, JB is funded by a studentship awarded by the Rosetrees Trust and AK is supported by a Leukaemia and Lymphoma Research studentship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Bullenkamp, J., Cole, D., Malik, F. et al. Human Gyrovirus Apoptin shows a similar subcellular distribution pattern and apoptosis induction as the chicken anaemia virus derived VP3/Apoptin. Cell Death Dis 3, e296 (2012). https://doi.org/10.1038/cddis.2012.34
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2012.34
Keywords
This article is cited by
-
Metagenomic characterisation of additional and novel avian viruses from Australian wild ducks
Scientific Reports (2020)
-
Novel characteristics of the avian gyrovirus 2 genome
Scientific Reports (2017)
-
VP2 of Chicken Anaemia Virus Interacts with Apoptin for Down-regulation of Apoptosis through De-phosphorylated Threonine 108 on Apoptin
Scientific Reports (2017)
-
Human Gyrovirus-Apoptin Interferes with the Cell Cycle and Induces G2/M Arrest Prior to Apoptosis
Archivum Immunologiae et Therapiae Experimentalis (2017)
-
Avian gyrovirus 2 in poultry, China, 2015–2016
Emerging Microbes & Infections (2016)