Abstract
The three major subgroups of the Bcl-2 family, including the prosurvival Bcl-2-like proteins, the proapoptotic Bcl-2 homology (BH)3-only proteins and Bax/Bak proteins, regulate the mitochondrial apoptotic pathway. In addition, some outliers within the Bcl-2 family do not fit into these subgroups. One of them, Bcl-G, has a BH2 and a BH3 region, and was proposed to trigger apoptosis. To investigate the physiological role of Bcl-G, we have inactivated the gene in the mouse and generated monoclonal antibodies to determine its expression. Although two isoforms of Bcl-G exist in human, only one is found in mice. mBcl-G is expressed in a range of epithelial as well as in dendritic cells. Loss of Bcl-G did not appear to affect any of these cell types. mBcl-G only binds weakly to prosurvival members of the Bcl-2 family, and in a manner that is independent of its BH3 domain. To understand what the physiological role of Bcl-G might be, we searched for Bcl-G-binding partners through immunoprecipitation/mass spectroscopy and yeast-two-hybrid screening. Although we did not uncover any Bcl-2 family member in these screens, we found that Bcl-G interacts specifically with proteins of the transport particle protein complex. We conclude that Bcl-G most probably does not function in the classical stress-induced apoptosis pathway, but rather has a role in protein trafficking inside the cell.
Similar content being viewed by others
Main
Proteins of the Bcl-2 family are divided into three subgroups according to their function and the presence of characteristic Bcl-2 homology (BH) domains. The prosurvival members (Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1/Bfl-1) contain four BH domains and are critical for cell survival. The proapoptotic proteins Bax and Bak, which also contain all four BH domains and are structurally similar to their prosurvival relatives, are essential for mitochondrial outer membrane permeabilisation. The BH3-only proteins (Bim, Bid, Puma, Bmf, Bik, Bad, Noxa and Hrk) are essential initiators of apoptosis that cause activation of Bax and Bak, either directly or indirectly by binding to and blocking the prosurvival Bcl-2 family members.1 The presence of the BH1, BH2 and BH3 domains in the multi-domain proteins determines their three-dimensional structure, in particular the existence on their surface of a groove that functions as a receptor for the BH3 domain of BH3-only or the Bax/Bak proteins. Mutations in the conserved residues of the BH3 domain of proapoptotic proteins almost invariably modify or abrogate their interaction with the groove of the prosurvival Bcl-2-like proteins.2
A number of other proteins harbouring a BH3-like domain (and sometimes another BH domain) have been described.3 Although many of these proteins have been included in the Bcl-2 family, their role in stress-induced apoptosis is often controversial, and some of these proteins have even been implicated in diverse non-apoptotic cellular functions, such as cell cycle regulation, DNA repair, ubiquitination, metabolism or autophagy.3
BCL-G, also known as BCL2L14, was first described as a novel proapoptotic member of the Bcl-2 family.4 The human BCL-G gene encodes two major isoforms, BCL-GL and BCL-GS. BCL-GL contains both BH3 and BH2 domains, whereas BCL-GS contains only the BH3 domain. BCL-GS was reported to kill cells by binding and neutralisation of prosurvival BCL-XL. In contrast, BCL-GL did not bind BCL-XL and showed only poor killing activity.4 Mouse Bcl-G is 68% identical and 78% similar to human BCL-GL.5 Functional characterisation of mBcl-G has been limited, and most studies have considered its proapoptotic nature as a fait accompli.
To characterise this unusual member of the Bcl-2 family, we have generated highly specific monoclonal antibodies (mAb) and Bcl-G-deficient mice. Our results highlight differences between the mouse and human Bcl-G genes and suggest that Bcl-G may not function as a classical BH3-only protein.
Results
Generation of Bcl-G-deficient mice
Targeting of the mouse Bcl-G gene in embryonic stem (ES) cells was achieved by introducing loxP sites flanking the ATG-containing exon 3 (Figure 1a). Hygromycin-resistant Bruce 4 clones were isolated and their genomic DNA analysed by Southern blotting (Figure 1b). Bcl-G gene-targeted mice were established and maintained on an inbred C57BL/6 genetic background, and were genotyped by PCR (Figure 1c). Bcl-G−/− mice were born at the expected Mendelian frequency from intercross matings of Bcl-G+/− mice (Figure 1d). The general appearance and behaviour of these mice was normal and they were indistinguishable from their wild-type (WT) littermates.
Targeting strategy and generation of Bcl-G knock-out mice. (a) Schematic diagram depicting the Bcl-G locus targeting strategy. Open boxes represent exons and grey boxes represent intronic DNA (corresponding region on WT locus is indicated by dashed lines). H denotes the hygromycin resistance selection cassette (frt/loxP/pGK-Hygro/frt), whereas triangles and rectangles indicate the loxP sites and frt sites, respectively. The locations of the 5′- and 3′-external probes used for Southern blot analysis are shown. (b) Southern blot analysis on ScaI-digested genomic DNA to identify homologous recombination at the Bcl-G locus. Correctly targeted ES cell clones show a 16.6 kb fragment with the 5′-external probe and a 6.8 kb fragment with the 3′-external probe. (c) Bcl-G−/− mice were identified using a three-primer genotyping PCR protocol on tail DNA. Wild-type mice show a single band of 202 bp, whereas Bcl-G−/− mice show a single band of 346 bp. Bcl-G+/− mice show both bands. (d) Genotypes of offspring produced from crosses of Bcl-G+/− mice. The actual numbers are shown in brackets
As expected, no RNA could be detected by RT-PCR in tissues from Bcl-G−/− mice (Figure 2a). Our mAb (clone 2E11) detected a 38-kDa band in WT testis, and this band was absent from Bcl-G−/− testis (Figure 2b). Interestingly, only one Bcl-G isoform is produced from the mouse Bcl-G gene, whereas a long (BCL-GL) and a short (BCL-GS) isoform are produced from human BCL-G.4 In the testis, Bcl-G immunoreactivity was limited to the late-stage spermatids within the seminiferous tubules (Figure 2c). No staining was seen in the mature sperm found in the epididymal tubules (data not shown). Interestingly, Bcl-G-deficient male (and female) mice exhibited normal reproductive behaviour and produced litters of normal size.
Loss of Bcl-G does not affect testicular development. (a) RT-PCR performed on mRNA using inter-exon primers confirmed the absence of functional Bcl-G transcripts in tissues of Bcl-G−/− mice. PCR using actin-specific primers served as a loading control. (b) Western blotting using Bcl-G-specific mAbs detected the presence of Bcl-G protein in testes of WT and Bcl-G+/− mice. No Bcl-G protein was detected in the testis from Bcl-G−/− mice. Membranes were reprobed for HSP70 as a loading control. (c) Anti-Bcl-G immunohistochemistry revealed that Bcl-G expression is limited to the late stage spermatids within the seminiferous tubules. Representative photomicrographs are shown at × 100 (bar represents 100 μm) and × 400 magnification (bar represents 40 μm)
Bcl-G is expressed in diverse mouse tissues but is not required for their development and function
In addition to the male reproductive organs, high levels of mBcl-G were also found in the thymus, small intestine and colon. Intermediate expression levels were seen in pancreas, spleen and lung, whereas little or no Bcl-G expression was observed in the brain, kidney and liver (Figure 3a). Mouse Bcl-G protein is most similar to human BCL-GL, and its expression closely matches what is known for hBCL-GL mRNA.4
Bcl-G is expressed in eptihelial cells in a wide range of tissues and in DCs in the haematopoietic organs. (a) Western blot analysis of organs from C57BL/6 (WT) mice to determine distribution of Bcl-G. cDCs indicate FACS-sorted splenic conventional DCs (CD11chi CD45RA−), whereas IECs denote IECs isolated from WT intestines. Lysates from testis of Bcl-G−/− mice were used as a negative control. The membranes were reprobed for HSP70 as a loading control. (b) Immunohistochemistry using specific mAbs revealed cell type-specific expression of Bcl-G in the colon, mammary gland, spleen and thymus. Tissue sections from Bcl-G−/− mice were used as negative controls. The bars represent 100 μm. (c) Bcl-G expression in the various splenic DC subsets was analysed by western blotting. pDC stands for plasmacytoid DC (CD11cint CD45RA+); CD8, CD4 and double negative (DN) indicates CD8+ CD4−, CD8− CD4+ and CD8− CD4− cells, respectively). Bcl-G content in DCs stimulated with CpG DNA for 24 h was also analysed by western blotting
Bcl-G expression was high in many epithelial cells, in particular those lining the gastrointestinal tract and the lumen of mammary ducts (Figure 3b and data not shown). However, no obvious defects were found in these organs in Bcl-G−/− mice. Bcl-G−/− intestines had similar lengths of villi and crypt depths as those from WT mice (Figure 3b). The numbers of Paneth and goblet cells were also normal, indicating that Bcl-G is not required for intestinal epithelial differentiation (data not shown). In the mammary gland, Bcl-G loss did not affect ductal morphogenesis, and the lumens of ducts in Bcl-G−/− females were clear and well developed (Figure 3b). As the survival and growth of pups from litters of Bcl-G−/− females was no different from those of WT mothers, we conclude that Bcl-G deficiency did not significantly affect mammary development during pregnancy, nor did it affect milk production or nursing behaviour (data not shown).
Bcl-G is expressed by dendritic cells
Bcl-G was expressed in large cells within the thymic medulla and the centre of lymphocytic follicles of the spleen and lymph nodes (Figure 3b, and data not shown). No significant Bcl-G staining was detected in lymphocytes. The Immunological Genome Project gene expression database revealed high expression of Bcl-G mRNA in CD8+ dendritic cells (DCs).6 We examined Bcl-G expression in select DC subsets from WT spleen using our Bcl-G-specific antibodies (Figure 3c). In accordance with the mRNA expression results, conventional DCs (cDCs; CD11chi CD45RA−) had significantly higher levels of Bcl-G than plasmacytoid DCs (pDCs; CD11cint CD45RA+) (Figure 3c). This high expression was largely contributed by the CD8+ cDC subset, although Bcl-G was also present at lower levels in the CD4+ and double negative cDC subsets (Figure 3c). Bcl-G expression was increased in splenic cDCs upon stimulation with CpG oligonucleotides, perhaps indicating a role for Bcl-G in the Toll-like receptor 9 signalling pathway (Figure 3c).
The thymic medulla contains epithelial cells that are present in close proximity to the thymic DCs. As Bcl-G displayed expression in certain epithelial cell types in other organs, we investigated whether it is present in medullary thymic epithelial cells. However, double immunofluorescence staining for Bcl-G and the medullary thymic epithelial cell marker keratin-5 on WT thymus sections did not show any colocalisation, demonstrating that the DCs must constitute the majority of the Bcl-G-expressing cells in the thymus (Supplementary Figure 1).
Bcl-G−/− mice have normal haematopoietic cell subset composition
To investigate the impact of Bcl-G loss on the haematopoietic compartment, we analysed the cellular composition of blood, thymus and spleen in adult Bcl-G-deficient mice (Figures 4a–c). Overall numbers of red blood cells, white blood cells and platelets were all normal in Bcl-G-deficient mice (Figure 4a and data not shown). Similarly, thymi and spleens of Bcl-G−/− mice had similar weights and total cellularities as those of WT mice (Supplementary Figure 2, and Figures 4b and c). Bcl-G−/− mice also had normal numbers of each of the major thymic and splenic subpopulations analysed, showing that loss of Bcl-G did not affect the development and homeostasis of the mouse haematopoietic system (Figures 4b and c).
Bcl-G−/− mice have normal haematopoietic cell subset composition, and Bcl-G deficiency does not protect splenic DCs from apoptosis. (a) Leukocyte cell subset composition of 8–12-week-old WT and Bcl-G−/− mice was determined using an ADVIA blood analyser. Single-cell suspensions made from (b) thymi and (c) spleens of these mice were analysed by FACS after staining with antibodies against various cell surface markers to determine the numbers of the indicated cell subpopulations. Splenic subsets determined included B220+sIgMhisIgDlo immature B and B220+sIgMlosIgDhi mature B cells, Mac-1+Gr-1− macrophages, Mac-1+Gr1+ granulocytes and Ter119+ nucleated erythroid cells. (d) Splenic DC subset composition was determined by flow cytometric analysis. The absolute numbers per spleen of cDC (CD11chiCD45RA−), pDC (CD11cintCD45RA+), CD8+ cDC (gated cDC, CD8+Sirpα−) and CD8− cDC (gated cDC, CD8−Sirpα+) are shown (see Supplementary Figure 3). (e) FACS-sorted splenic DC subpopulations from WT and Bcl-G−/− mice were plated into 96-well plates with or without addition of the DC survival promoting cytokine GM-CSF. Cell viability was determined at specific time points using propidium iodide staining followed by flow cytometric analysis. Data are presented as percentages of PI− live cells versus time. The results shown represent at least three independent experiments (n=24 of each genotype). Error bars indicate S.E.M.
Bcl-G is dispensable for splenic DC development and apoptosis
As Bcl-G is highly expressed in DCs, particularly the CD8+ subtype, we next measured the numbers of the various splenic DC subsets in Bcl-G−/− mice using flow cytometry (Figure 4d and Supplementary Figure 3). The numbers of Bcl-G−/− DCs were comparable to those in WT mice (Figure 4d and Supplementary Figure 3). Because of the higher expression of Bcl-G in CD8+ DCs relative to other DC subsets, we also examined the function of the Bcl-G−/− CD8+ DCs, using several antigen cross-presentation assays (Supplementary Figure 4). However, neither the in vivo cross-presentation assay nor the CTL killing assay revealed any difference between the WT and Bcl-G-deficient DCs, indicating that Bcl-G may not be required for cross-presentation and naive T-cell activation (Supplementary Figure 4).
We next compared the survival of Bcl-G−/− and WT DCs in culture (Figure 4e). Splenic DCs from the three subsets (pDC, CD8+ and CD8− cDC) were purified by FACS and placed in medium with or without the survival factor granulocyte-macrophage colony-stimulating factor (GM-CSF; 10 ng/ml). Consistent with previous reports,7, 8 WT DCs, particularly the pDC subtype, underwent rapid cell death in vitro, with only ∼20% pDCs surviving after 24 h in culture (Figure 4e). There was no significant difference in the rate of death of Bcl-G−/− DCs from any of the three subtypes when compared with their WT counterparts (Figure 4e). These results demonstrate that Bcl-G is not required for spontaneous DC apoptosis, both in the presence or absence of GM-CSF. Transient overexpression of mouse Bcl-G did not cause significant killing of 293T cells (Supplementary Figure 5A), further indicating that mBcl-G lacks significant pro-apoptotic activity.
Bcl-G does not bind strongly to the prosurvival Bcl-2-like proteins
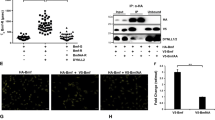
The complex interactions between three major subgroups of the Bcl-2 family determine whether apoptosis is activated or not.9, 10 Human BCL-GS was reported to bind BCL-XL through its BH3 domain, whereas the presence of the additional BH2 domain in hBCL-GL was suggested to prevent its binding to BCL-XL.4 As mBcl-G is most similar to hBCL-GL, and as we could not find any evidence that mBcl-G exerted significant proapoptotic activity, we investigated whether it could even bind to the antiapoptotic Bcl-2 family members or Bax/Bak. Haemagglutinin (HA)-tagged mBcl-G was transiently overexpressed together with Flag-tagged prosurvival proteins (Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1), Flag-Bax, Flag-Bak or empty vector as a negative control (Figure 5a). Bcl-G could bind weakly to all five prosurvival Bcl-2 family members when they were overexpressed (Figure 5a and Supplementary Figure 5B). In contrast, there was no evidence for significant interaction between mBcl-G and Bax or Bak (Figure 5a).
Weak interaction of Bcl-G with prosurvival Bcl-2 family members, which does not require its BH3 domain. Mammalian expression vectors encoding HA- and Flag-tagged proteins were co-transfected into 293T cells: (a) Full-length or (b) mutant HA-mBcl-G, together with Flag-tagged prosurvival proteins; (c) HA-mBimS (WT or mutant BH3) with Flag-Bcl-xL. Empty HA and Flag vectors were used as negative controls (−). Immunoprecipitations were carried out using Protein G sepharose beads and antibodies against HA, Flag or Glu-glu (negative control). Immunoprecipitates were separated on SDS-PAGE gels and tested for the presence of co-immunoprecipitated Flag- or HA-tagged proteins by western blotting
To determine the role of the BH3 domain in this weak binding, we generated several mutants of mBcl-G, including one lacking the entire BH3 domain (ΔBH3), one in which the conserved leucine within the BH3 domain was changed to alanine (L217A) and one which had the conserved glutamine and aspartate residues mutated to alanine (Figure 5b). Unexpectedly, these mutant Bcl-G proteins retained the ability to interact weakly with Bcl-xL and all four other prosurvival proteins (Figure 5b and data not shown). These results indicate that Bcl-G might not bind to the prosurvival Bcl-2 family members in the same manner as the canonical BH3-only proteins, such as Bim or Puma. In accordance with what was reported for hBCL-GL,4 removal of the BH2 domain from mBcl-G increased its binding to Bcl-xL (Figure 5b and Supplementary Figure 5B).
To further examine the ability of the BH3 domain of mBcl-G to bind to the prosurvival Bcl-2 family members, we generated a mutant form of BimS in which the BH3 domain was replaced with the BH3 domain of mBcl-G, and tested its binding to Bcl-xL (Figure 5c). This experiment revealed that BimS(Bcl-G BH3), such as BimS(4E), a well-described non-Bcl-xL-binding mutant of BimS,11 did not bind Bcl-xL (Figure 5c). This indicated that the BH3 domain of Bcl-G might be incapable of binding to the prosurvival Bcl-2 family members. Indeed, replacement of the BH3 domain of Bim with that of Bcl-G also resulted in complete loss of Bim’s killing potential when overexpressed in 293T cells (Supplementary Figure 5A). Finally, we were unable to observe any endogenous binding of Bcl-G to Bcl-xL, Bcl-2 and Mcl-1 in mouse tissue (testis, thymus or spleen) protein lysates when we immunoprecipitated either Bcl-G or Bcl-xL (Supplementary Figure 6). Collectively, these observations suggest that Bcl-G might not function as a classical BH3-only protein.
Novel potential interacting partners for Bcl-G
We then conducted a proteomic screen to identify potential new interacting partners for Bcl-G and thus gain clues on a possible alternative function. The strategy is depicted in Figure 6. Whole-cell lysates were prepared from intestinal epithelial cells (IECs) isolated from WT or Bcl-G−/− mice. The presence of Bcl-G in the lysates from the WT cells but not in those from the control (Bcl-G−/−) cells was verified by western blotting (Supplementary Figure 7). Bcl-G protein complexes were immunoprecipitated from these protein lysates using two different Bcl-G-specific mAbs, 2E11 and 10C9, that recognise different epitopes within the Bcl-G protein (Figure 6). Immunoprecipitates were analysed by mass spectrometry.
Identification of Bcl-G binding partners using IP–MS proteomic screening. Protein lysates made from IECs of WT or Bcl-G−/− mice (1) were used to immunoprecipitate Bcl-G using two independent anti-Bcl-G mAbs, 2E11 or 10C9 (2). Immunoprecipitated proteins were resolved by SDS-PAGE and visualised by Coomassie brilliant blue staining (3). Asterisks indicate the Bcl-G bands present in lane 1 (WT cell lysate, mAb 2E11) but not lane 2 (Bcl-G−/− cell lysate, mAb 2E11). Positions of molecular weight markers (MW) are shown. Corresponding regions for each sample were excised, trypsin-digested and sequenced by mass spectrometry (4). The 326 proteins identified in the screen were classified by their presence in each sample. Non-specific proteins were identified by their presence in either or both of the pull-downs from Bcl-G−/− cell lysates (216 proteins). Potential Bcl-G-interacting partners were identified as being present in either or both of the WT samples and absent in the Bcl-G−/− samples (110 proteins)
A total of 327 unique proteins were identified in this screen (Figure 6). Of these, 216 proteins could be eliminated as non-specific interactors, as they were found in all the samples, including immunoprecipitates from Bcl-G−/− cells. As expected from the western blots, Bcl-G-derived peptides were only found in the WT samples (Supplementary Figure 8). A total of 110 unique proteins that potentially interact with Bcl-G were identified (Figure 6 and Supplementary Figure 9). Of these, 19 proteins were found in immunoprecipitates with both antibodies (Figure 6 and Supplementary Figure 9D). Notably, none of the Bcl-2 family members were identified in this screen, consistent with our overexpression/co-immunoprecipitation studies (Figure 5).
Yeast two-hybrid screening identifies Trappc6b as a Bcl-G-binding protein
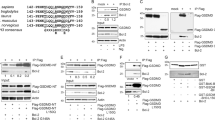
We reasoned that an independent approach would help to validate and strengthen the results of the IP–MS experiment. We therefore used full-length Bcl-G as a bait to perform a yeast two-hybrid (Y2H) screening of a normalised mouse universal cDNA library (Clontech, Mountain View, CA, USA). In a screen of approximately 3 × 106 transformants, we obtained 52 colonies growing on plates containing selective medium. Of the cDNAs picked up in this screen, three were in the correct reading frame. cDNAs encoding the transport particle protein (TRAPP) complex 6b (Trappc6b) represented 19 of the 52 isolated clones. The other clones encoded parts of Sorting nexin 14 (Snx14) and Transmembrane protein 167B (Tmem167B) and were each found once. Importantly, none of the Bcl-2 family members were pulled down as potential Bcl-G-binding partners in this screen. In addition, mBcl-G did not appear to bind to itself when overexpressed, in contrast to a previous report.12
Out of the three potential hits, we focused our attention on Trappc6b, as three components of TRAPP complexes (Trappc3, Trappc4 and Trappc5) had been identified in the IP–MS experiment (Table 1). To confirm the Y2H screening results, we introduced the pGADT7–Trappc6b plasmid into the AH109 yeast strain together with bait plasmid pGBT9–Bcl-G (FL) or pGBT9 vector alone (V; Figure 7a). Only yeast cells transformed with both Trappc6b, and Bcl-G survived on -Leu-Trp-His-Ade plates and turned blue in the X-gal assay that detects the presence of α-galactosidase activity (Figure 7a). This interaction was specific, as deleting particular regions (amino acids 103–136 and 273–328) of the Bcl-G protein resulted in lack of α-galactosidase activity and no growth on the selective plates (Figure 7a and Supplementary Figure 10). In a pull-down experiment, ectopically expressed Flag–Trappc6b was found to bind to His-tagged Bcl-G, immobilised on Ni-NTA agarose beads (Figure 7b).
Trappc6b interacts with mBcl-G in the Y2H assay. (a) pGBT9-DBD empty vector or pGBT9-DBD-mBcl-G (full-length and truncation mutants M2–M10) were co-transformed with pGADT7-AD-Trappc6b into the yeast strain AH109. Positive interactions were tested by streaking transformants onto a -Leu-Trp-His-Ade plate (left) and the β-galactosidase assay was performed using X-gal staining of yeasts growing on a -Leu-Trp plate (right panel). The amino acid positions of the truncations within the Bcl-G mutants are shown in the schematic below. See Supplementary Figure 9 for more details. (b) Cell lysates made from 293T cells overexpressing Flag-Trappc6b were added to Ni-NTA beads pre-incubated with recombinant His-mBcl-G purified from Escherichia coli. Flag-Trappc6b bound to immobilised Bcl-G was identified by anti-Flag western blotting. Detection of unbound Trappc6b in the control sample indicates input. Probing for HSP70 served as a control that equal amounts of protein lysates were used
Collectively, the results from the Y2H screen and the IP–MS experiment confirm that Bcl-G does not bind significantly to other members of the Bcl-2 family and strongly suggest that it is involved in vesicle trafficking and protein transport across the cells by interacting with the TRAPP complex.
Discussion
Proteins of the Bcl-2 family are characterised by the presence of BH domains that are important in determining the three-dimensional structure of these proteins and their interactions with the other members of the family. In this regard, mBcl-G and its human homologue BCL-GL are rather unique members of the Bcl-2 family, as they contain both BH3 and BH2 domains.4, 5 The only other protein in this family to contain such a combination of BH domains is Bfk.13 Overexpression of hBCL-Gs was found to induce apoptosis in certain cell lines, whereas hBCL-GL did not.4 Accordingly, BCL-G is widely (if not universally) considered to encode yet another proapoptotic BH3-only protein, but this proapoptotic activity has not been confirmed in an independent study, let alone within a physiological context. A database search indicated that the BCL-GS isoform only exists in humans, as the alternative splice acceptor site that is used to produce it is not conserved in other species. In accord with its lack of killing activity (even when overexpressed), hBCL-GL did not bind BCL-XL.4 Although we detected a weak interaction of mBcl-G with several prosurvival Bcl-2 family members, this interaction was still observed when the BH3 domain of Bcl-G was mutated or removed. In fact, closer examination indicates that the BH3 domain of Bcl-G does not conform to the BH3 consensus sequence; for example, the uniformly hydrophobic residue in position Φ3 is replaced with a serine (S220). This may explain why mBcl-G binds poorly to prosurvival family members. Strikingly, no Bcl-2 family member, prosurvival or proapoptotic, was pulled down in either our proteomic screen or the Y2H screen. Collectively, these results indicate that mBcl-G is most likely not involved in interactions with other Bcl-2 family members and, therefore, probably does not function in the intrinsic apoptotic pathway.
To determine which tissues and cell types normally express Bcl-G, we have generated three highly specific mAbs.14 In western blots, mBcl-G appears as a ∼38-kDa band that is not present in Bcl-G−/− animals. It is worth noting that some of the earliest commercially available anti-BCL-G antibodies detect a single ∼22-kDa band that corresponds neither to the size of hBCL-GL nor that of hBCL-GS. We found that Bcl-G is highly expressed in many epithelial cells, as well as in DCs. This guided the choice of the source of protein lysates for our IP–MS experiment, as we wanted to avoid using Bcl-G overexpression. Our search for novel interacting partners using two independent approaches led to the discovery that Bcl-G may be a component of the TRAPP complex, which is involved in vesicle-mediated protein transport. Two large multi-subunit TRAPP complexes have been identified in yeast, whereas there seems to be only one TRAPP complex in mammals.15 These complexes function in the endoplasmic reticulum (ER) to Golgi transport, as well as intra-Golgi transport. Our immunofluorescence studies did not reveal any specific subcellular localisation of Bcl-G in the Golgi, ER or within vesicular structures when overexpressed in HeLa cells.14 However, Bcl-G overexpressed in HeLa cells may not reflect its physiological localisation as it was shown in the case of Trapp complex subunit 3.16 As Bcl-G does not exist in yeast, it is difficult to predict what its precise function in the TRAPP complex is, and more work is required to determine this function.
The BCL-G gene is located on human chromosome region 12p12–13, which has been associated with a range of malignancies.17 A search for a putative prostate cancer tumour suppressor gene within this region revealed that Bcl-G transcript levels were downregulated in tumour samples compared with normal prostate tissue.18 No mutations or expression changes of the Bcl-G gene were found in children with acute lymphoblastic leukaemia,17 but a TEL–AML1 fusion transcript involving BCL-G was reported in another.19 In breast cancer, Bcl-G expression was found to be abnormally reduced and possibly regulated post-translationally by maternal embryonic leucine zipper kinase, a recently identified protein kinase and candidate oncoprotein upregulated in several types of cancer.20 In all these studies, BCL-G was considered a proapoptotic BH3-only protein and, as such, a potential tumour suppressor. These considerations must now be revised in the light of our results, which show that Bcl-G is not an inducer of apoptosis, although they do not preclude by any means that BCL-G may still work as a tumour suppressor. Hence, the importance of Bcl-G in initiation and progression of cancer remains to be studied.
Certain cell types with prominent Bcl-G expression were studied in detail in an attempt to identify a defect caused by the loss of this protein. Most DCs have a short lifespan, and upon activation and antigen presentation, they rapidly undergo apoptosis to limit the potency of the immune response that had been elicited.8, 21 Members of the Bcl-2 protein family have critical roles in DC apoptosis.21, 22, 23 For example, DCs from Bim−/− mice are abnormally resistant to spontaneous apoptosis, leading to enhanced T-cell activation and autoantibody production in vivo.7, 24 In contrast, loss of Bcl-G did not impact on DC lifespan, nor did it significantly impact on their main functions, such as antigen cross presentation.
The lack of an obvious phenotype in Bcl-G-deficient mice could be attributed to the existence of functional redundancy, as was reported for other members of the Bcl-2 protein family.25 However, our results have indicated that Bcl-G might not have a proapoptotic function after all. It thus seems reasonable to investigate the function of Bcl-G further before embarking on time-consuming and costly double-knockout studies.
Several BH3 domain-containing proteins have been shown to function outside the conventional Bcl-2-regulated apoptotic pathway, including cell cycle control, autophagy, calcium homeostasis and ER morphogenesis.3 It appears likely that Bcl-G may have to be added to this list. The Bcl-G-deficient mice as well as the mAbs that we have generated should prove valuable tools to further study this intriguing Bcl-2 family member.
Materials and Methods
Generation of Bcl-G−/− mice
The targeting construct was designed to introduce loxP sites on either side of the ATG start codon containing exon 3, as well as an FRT-flanked PGK-hygromycin resistance cassette for screening purposes. The targeting construct was electroporated into C57BL/6-derived Bruce-4 ES cells.26 Homologous recombination events were identified by Southern blotting, using external 5′- and 3′-genomic probes, and blotting with a hygro-specific probe confirmed single-construct integration. Two correctly targeted ES cell clones were injected into blastocysts, resulting in two independent Bcl-G gene-targeted mouse strains. Exon 3 and the hygromycin-resistance cassette were deleted by crossing the resultant Bcl-GloxP/wt heterozygous mice with C57BL/6 cre deleter mice, and the Cre transgene was subsequently eliminated by crossing offspring to C57BL/6 mice. All of the mice analysed were devoid of hygro and cre. Genotyping of Bcl-G-deficient mice was performed using a three-primer PCR reaction using a common forward primer (5′-TCCGTCCCATTATAACCCTA-3′), together with two reverse primers (5′-CTGGAACGACAGAGGGCAAA-3′ and 5′-TTACCATCACAGAGCCAGAA-3′). The WT and mutant alleles produced products of 202 and 346 bp, respectively.
C57BL/6, B6.CH-2bm 1 (bm1) and OT-I27 mice were obtained from WEHI Bioservices (Kew, VIC, Australia). All animal procedures were reviewed and approved by the Melbourne Directorate Animal Ethics Committee.
RT-PCR analysis
RNA extraction was performed with the TRIzol method (Life Technologies, Mulgrave, VIC, Australia) according to the manufacturer’s instructions. cDNA was synthesised using the TaqMan Gold RT-PCR kit (Life Technologies). The PCR amplification step used GoTaq Green Master Mix (Promega, Alexandria, NSW, Australia) and 25 pmol of each forward (5′-CCCAAGCTCTCCAGAACAAG-3′) and reverse (5′-CAGCAGCTCAACAATCTTGC-3′) primers.
Western blotting and immunoprecipitation
HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, transiently transfected using Fugene 6 reagent (Roche, Dee Why, NSW, Australia), and were harvested 40 h post-transfection. Cells were lysed in 20 mM Tris-pH 7.4, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol and 1% Triton X-100 supplemented with complete protease inhibitor cocktail tablets (Roche). Lysates were incubated with protein G-sepharose beads and antibodies against HA (3F10; Roche), Flag (M2; Sigma-Aldrich, Rowville, VIC, Australia) or EE (Glu-Glu; Covance, North Ryde, NSW, Australia). Protein complexes were resolved on 4–12% Bis-Tris SDS-PAGE gels (Life Technologies) and detected on western blots with rat mAbs to Bcl-G (2E11)14 or HA and mouse mAbs to Flag (M2, Sigma-Aldrich), HSP70 (N6; a kind gift from Dr R Anderson, Peter MacCallum Cancer Research Centre, Melbourne, VIC, Australia) or Actin (AC-40; Sigma-Aldrich). Labelled protein bands were detected by incubation with goat anti-mouse Ig or goat anti-rat IgG antibodies conjugated to horseradish peroxidase (Southern Biotech, Birmingham, AL, USA), followed by chemiluminescence analysis (ECL; GE Healthcare, Rydalmere, NSW, Australia)
Histology and tissue immunohistochemistry
Mouse tissues were fixed in 10% formalin. Protocols used for immunohistochemistry and immunofluorescence have been described.14
Immunofluorescence and flow cytometric analysis
Peripheral blood was analysed with the ADVIA 120 Hematology System (Bayer, Tarrytown, NY, USA). Single-cell suspensions prepared from thymi and spleens were counted using the CASY Cell Counter (Roche). Cells were stained for flow cytometric analysis using fluorochrome (fluorescein isothiocyanate, PE, APC or PerCP)-conjugated antibodies to Thy1.2 (T24.31.2), CD8 (YTS169), B220 (RA3-6B2), IgM (5.1), IgD (1126C), Gr-1 (8C5), CD3 (KT3-1.1), CD4 (YTA-321), CD11b (Mac-1) and Ter-119 (erythroid cell marker). Staining with propidium iodide (5 μg/ml) was used to label dead cells. Data were acquired on either a FACScan or an LSR II flow cytometer (BD Biosciences, San Jose, CA, USA). All data analysis was performed using FlowJo software (TreeStar, Ashland, OR, USA).
DC isolation and treatments
DCs were isolated from spleens of mice according to previously published protocols,28 followed in some cases by high-speed cell sorting (MoFlo, Cytomation, Fort Collins, CO, USA). DC populations purified from WT or Bcl-G−/− mice were added to 96-well plates at 1 × 105 cells per well in RPMI medium 1640 supplemented with 10% FCS, 50 μM 2-mercaptoethanol, 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin with or without supplementation with 100 ng/ml GM-CSF (PeproTech, Rocky Hill, NJ, USA) and/or 0.5 μM CpG1168 (Geneworks, Hindmarsh SA, Australia). In vivo OT-I TCR transgenic T-cell proliferation assays and in vivo CTL assays were performed as described.29
Isolation of primary small IECs
Small intestines from either WT or Bcl-G−/− mice were removed, opened longitudinally, cut into 0.5-cm pieces and placed into ice-cold DMEM media containing 1 mM DTT. Gut pieces were shaken gently for 15 min at 37 °C, the dislodged cells filtered out and solid material placed in PBS solution containing 15 mM EDTA, followed by a further 10-min incubation with shaking at 37 °C. After vortexing to dislodge IECs, cell suspensions were passed through cell strainers (100 μm, BD Biosciences) and centrifuged at 4 °C at 300 × g for 5 min.
Mass spectrometry
Immunoprecipitates were resolved on a 4–12% Bis-Tris SDS-PAGE gel (Invitrogen, Grand Island, NY, USA) and visualised by staining with the Colloidal Blue Staining Kit (Invitrogen). The gel regions between 25–55 kDa were excised and divided into eight equal slices, which were then subjected to in-gel reduction, alkylation and tryptic digestion. Extracted peptides were injected and fractionated by nanoflow reverse-phase liquid chromatography on a nano LC system (1200 series; Agilent, Santa Clara, CA, USA). The nano HPLC was coupled online to an LTQ-Orbitrap mass spectrometer equipped with a nanoelectrospray ion source (Thermo Fisher Scientific, Scoresby, VIC, Australia) for automated MS/MS. For protein identification within gel samples, LC-MS/MS data were searched against a non-redundant protein decoy database comprising sequences from the latest version of LudwigNR (Mouse Species, Melbourne, VIC, Australia). Mass spectra peak lists were extracted using extract-msn as part of Bioworks 3.3.1 (Thermo Fisher Scientific) linked into Mascot Daemon (Matrix Science, London, UK). Peak lists for each nano-LC-MS/MS run were used to search MASCOT v2.2.04 search algorithm (Matrix Science) provided by the Australian Proteomics Computational Facility (www.apcf.edu.au).
Y2H assay
A pre-transformed normalised Matchmaker mouse universal cDNA library prepared with pGADT7-RecAB was purchased from Clontech and screened by the Matchmaker GAL4 Two-Hybrid System (Clontech) according to the manufacturer’s protocol. Full-length mBcl-G cDNA fragment was amplified by PCR and cloned into pGBT9 (Clontech). The yeast Saccharomyces cerevisiae strain AH109 (maintained in YEPD medium), which secretes α-galactosidase under the control of the MEL1 region, was transformed with pGBT9-mBcl-G and grown on a medium lacking tryptophan. The clone including the bait plasmid was transformed with the library plasmids. The transformed yeast cells were grown on 2% agar plates of a dropout medium lacking tryptophan, leucine, histidine and adenine (-Leu-Trp-His-Ade). The resulting colonies grown on the drop-out plate were inoculated again on a new drop-out plate containing 20 μg/ml X-α-Gal (5-bromo-4-chloro-3-indolyl-α-O-galactopyranoside; Clontech) and incubated at 30 °C for 7 days. Yeast co-transformed with pGADT7 and pGBT9 empty vectors, or pGBT9-p53 and pGADT7-LgT were used, respectively, as negative and positive controls. Total DNA was prepared from all positive blue colonies and introduced into E. coli TOP10 competent cells (Invitrogen). The prey plasmids were recovered and sequenced.
Ni–NTA agarose purification
Forty-eight hours post-transfection, 293T cells were washed with PBS and resuspended in onyx lysis buffer. Ni–NTA agarose beads (20 μl) (Qiagen, Chadstone, VIC, Australia) pre-incubated for 1 h with recombinant His-Bcl-G protein (12 μg) were washed, added to cell extracts and rotated at 4 °C for 2 h. Precipitates were washed four times with lysis buffer and then boiled in 40 μl 2 × NuPAGE LDS buffer (Invitrogen) followed by SDS-PAGE analysis.
Data analysis
All graphing and statistical analyses were performed using the Prism v4.0 (GraphPad Software, San Diego, CA, USA).
Abbreviations
- BH:
-
Bcl-2 homology domain
- cDC:
-
conventional dendritic cell
- pDC:
-
plasmacytoid dendritic cell
- HA:
-
haemagglutinin
- mAb:
-
monoclonal antibody
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- Y2H:
-
yeast two-hybrid
- TRAPP:
-
transport particle protein
References
Giam M, Huang DC, Bouillet P . BH3-only proteins and their roles in programmed cell death. Oncogene 2008; 27: S128–S136.
Hinds MG, Day CL . Regulation of apoptosis: uncovering the binding determinants. Curr Opin Struct Biol 2005; 15: 690–699.
Lomonosova E, Chinnadurai G . BH3-only proteins in apoptosis and beyond: an overview. Oncogene 2008; 27: S2–S19.
Guo B, Godzik A, Reed JC . Bcl-G, a novel pro-apoptotic member of the Bcl-2 family. J Biol Chem 2001; 276: 2780–2785.
Nakamura M, Tanigawa Y . Characterization of ubiquitin-like polypeptide acceptor protein, a novel pro-apoptotic member of the Bcl2 family. Eur J Biochem 2003; 270: 4052–4058.
Heng TS, Painter MW . The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008; 9: 1091–1094.
Chen M, Huang L, Wang J . Deficiency of Bim in dendritic cells contributes to overactivation of lymphocytes and autoimmunity. Blood 2007; 109: 4360–4367.
Chen M, Huang L, Shabier Z, Wang J . Regulation of the lifespan in dendritic cell subsets. Mol Immunol 2007; 44: 2558–2565.
Youle RJ, Strasser A . The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59.
Strasser A, Cory S, Adams JM . Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J 2011; 30: 3667–3683.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG et al. Differential targeting of pro-survival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17: 393–403.
Liu X, Pan Z, Zhang L, Sun Q, Wan J, Tian C et al. JAB1 accelerates mitochondrial apoptosis by interaction with proapoptotic BclGs. Cell Signal 2008; 20: 230–240.
Coultas L, Pellegrini M, Visvader JE, Lindeman GJ, Chen L, Adams JM et al. Bfk: a novel weakly proapoptotic member of the Bcl-2 protein family with a BH3 and a BH2 region. Cell Death Differ 2003; 10: 185–192.
Giam M, Mintern JD, Rautureau GJP, Hinds MG, Strasser A, Bouillet P . Detection of Bcl-2 family member Bcl-G in mouse tissues using new monoclonal antibodies. Cell Death and Disease 2012; 3: e-pub ahead of print 23 August 2012; doi:10.1038/cddis.2012.117.
Sacher M, Kim YG, Lavie A, Oh BH, Segev N . The TRAPP complex: insights into its architecture and function. Traffic 2008; 9: 2032–2042.
Yu S, Satoh A, Pypaert M, Mullen K, Hay JC, Ferro-Novick S . mBet3p is required for homotypic COPII vesicle tethering in mammalian cells. J Cell Biol 2006; 174: 359–368.
Montpetit A, Larose J, Boily G, Langlois S, Trudel N, Sinnett D . Mutational and expression analysis of the chromosome 12p candidate tumor suppressor genes in pre-B acute lymphoblastic leukemia. Leukemia 2004; 18: 1499–1504.
Latil A, Bieche I, Chene L, Laurendeau I, Berthon P, Cussenot O et al. Gene expression profiling in clinically localized prostate cancer: a four-gene expression model predicts clinical behavior. Clin Cancer Res 2003; 9: 5477–5485.
Abdelhaleem M, Yi Q, Beimnet K, Hitzler J . A novel TEL-AML1 fusion transcript involving the pro-apoptotic gene BCL-G in pediatric precursor B acute lymphoblastic leukemia. Leukemia 2006; 20: 1294.
Lin ML, Park JH, Nishidate T, Nakamura Y, Katagiri T . Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer Res 2007; 9: R17.
Kamath AT, Henri S, Battye F, Tough DF, Shortman K . Developmental kinetics and lifespan of dendritic cells in mouse lymphoid organs. Blood 2002; 100: 1734–1741.
Nopora A, Brocker T . Bcl-2 controls dendritic cell longevity in vivo. J Immunol 2002; 169: 3006–3014.
Hon H, Rucker EB, Hennighausen L, Jacob J . bcl-xL is critical for dendritic cell survival in vivo. J Immunol 2004; 173: 4425–4432.
Fuertes Marraco SA, Scott CL, Bouillet P, Ives A, Masina S, Vremec D et al. Type I interferon drives dendritic cell apoptosis via multiple BH3-only proteins following activation by PolyIC in vivo. PLoS One 2011; 6: e20189.
Coultas L, Bouillet P, Loveland KL, Meachem S, Perlman H, Adams JM et al. Concomitant loss of proapoptotic BH3-only Bcl-2 antagonists Bik and Bim arrests spermatogenesis. Embo J 2005; 24: 3963–3973.
Lemckert FA, Sedgwick JD, Korner H . Gene targeting in C57BL/6 ES cells. Successful germ line transmission using recipient BALB/c blastocysts developmentally matured in vitro. Nucleic Acids Res 1997; 25: 917–918.
Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR . T cell receptor antagonist peptides induce positive selection. Cell 1994; 76: 17–27.
Vremec D, Pooley J, Hochrein H, Wu L, Shortman K . CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J Immunol 2000; 164: 2978–2986.
Wilson NS, Behrens GM, Lundie RJ, Smith CM, Waithman J, Young L et al. Systemic activation of dendritic cells by Toll-like receptor ligands or malaria infection impairs cross-presentation and antiviral immunity. Nat Immunol 2006; 7: 165–172.
Acknowledgements
We thank M Robati for technical assistance; LA O’Reilly for sharing reagents and expertise; G Siciliano, J Coughlin, E Sutherland and S O’Connor for mouse care; J Corbin for automated blood analysis; B Helbert and C Young for genotyping; K Wycherley for mAb production; E Tsui, S Mihajlovic and their team for histological preparations; and H Patsiouras and E Kapp from the Joint Proteomics Laboratory for mass spectrometry analysis. This work was supported by the Australian NHMRC (Program Grant 461221, Independent Research Institutes Infrastructure Support Scheme Grant 361646, and a Career Development Award), the Leukemia and Lymphoma Society (Specialised Center of Research Grant 7015), and infrastructure support from the NHMRC (IRISS) and the Victorian State Government (OIS). M Giam was funded by University of Melbourne under the Endeavour IPRS scholarship programme. JD Mintern is the recipient of an NHMRC Career Development Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary information accompanies the paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Giam, M., Okamoto, T., Mintern, J. et al. Bcl-2 family member Bcl-G is not a proapoptotic protein. Cell Death Dis 3, e404 (2012). https://doi.org/10.1038/cddis.2012.130
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2012.130
Keywords
This article is cited by
-
BCL-G: 20 years of research on a non-typical protein from the BCL-2 family
Cell Death & Differentiation (2023)
-
Human BCL-G regulates secretion of inflammatory chemokines but is dispensable for induction of apoptosis by IFN-γ and TNF-α in intestinal epithelial cells
Cell Death & Disease (2020)
-
Loss of Bcl-G, a Bcl-2 family member, augments the development of inflammation-associated colorectal cancer
Cell Death & Differentiation (2020)
-
An RNA interference screen identifies new avenues for nephroprotection
Cell Death & Differentiation (2016)
-
MELK—a conserved kinase: functions, signaling, cancer, and controversy
Clinical and Translational Medicine (2015)