Abstract
To determine the role of biomarkers in the clinical management of respiratory complications (RC) in hematopoietic stem cell transplantation (HSCT) recipients, we have prospectively evaluated a cohort of 175 patients followed-up for 1 year after HSCT. To avoid misinterpretation, we have excluded both unidentified respiratory infections (RI) and mixed RI. A total of 64 RC were included. Plasma levels of C-reactive protein (CRP), procalcitonin (PCT) and proadrenomedullin (proADM) were measured at diagnosis and on day 3 and 7. Different cytokines were evaluated in serum on the first day. No HSCT recipients without RC were included as a control group. Compared with RI, non-infectious RC showed a significant increase in CRP, proADM and interleukin 6 on day 0 (P=0.005; P=0.03 and P=0.04, respectively). When only RI were considered, we observed that bacterial–fungal PI showed higher levels of CRP (P=0.02), PCT (P=0.04) and proADM (P<0.01). Persistent low levels of proADM biomarkers suggest viral infection (specificity and positive predictive value 100%). Patients dying of RC had PCT and proADM levels higher than survivors (P=0.002 and P=0.03, respectively). In HSCT recipients biomarkers increase in both infectious and non-infectious RC. They may have utility in the assessment of the severity of RC and in suspecting a viral etiology.
Similar content being viewed by others
Introduction
Respiratory complications (RC) are common and often a major cause of mortality in hematopoietic stem cell transplantation (HSCT) recipients.1, 2, 3, 4 In a recent series published by our group, almost 30% of patients undergoing an HSCT suffered at least one significant RC in the first year after transplantation, with an associated mortality of 23%.5 In these patients, obtaining an early diagnosis is essential for prescribing a specific treatment that may improve prognosis.6 However, this is a difficult task as the range of potential complications, both infectious and non-infectious, is considerable and the clinical and radiologic presentation is often similar. These RC are thus often treated with different empirical drugs and often unnecessary, potentially toxic and expensive therapies, which can lead to antimicrobial resistance. The availability of new biomarkers with high sensitivity and specificity for infections may facilitate the clinical management of RC in HSCT recipients. C-reactive protein (CRP) is an acute phase protein produced by the liver, which has been widely used as a biochemical inflammatory marker.7, 8 More recently, researchers have been focussing on procalcitonin (PCT) and proadrenomedullin (proADM), two peptides derived from the calcitonin gene family, with distinct functions and properties.9, 10, 11 Although these biomarkers have proved useful for the management of systemic infections in immunocompetent patients, their value in HSCT recipients is still the subject of debate.12, 13 Thus, it has been shown that patients with GvHD may exhibit PCT positivity. Furthermore, different inflammatory non-infectious RC related to transplantation may cause an increase in different cytokines and inflammatory mediators, potentially diminishing the usefulness of biomarkers in clinical practice.14 Most series published in immunocompromised patients are retrospective and include a heterogeneous population of oncologic patients. Moreover, the episodes evaluated do not always have a specific etiology or do not have a pulmonary origin.15, 16 Finally, most series do not include a control group of HSCT recipients without RC. Although we neither have a control group, in this study we have tried to circumvent other aforementioned shortcomings. Thus, our cohort of HSCT recipients has been prospectively evaluated for one year.5 Also, only respiratory infections (RI) with an identified microorganism have been included. Finally, to avoid confounding information, mixed RI, a common occurrence in HSCT recipients, have been excluded.5
The aim of this study was to determine the role of different relevant biomarkers in identifying the type and severity of RC in HSCT recipients.
Materials and methods
Patients and transplantation procedure
The information provided in this manuscript comes from a prospective interventional study to evaluate RC in a cohort of consecutive HSCT recipients followed-up for 1 year. Details of this protocol have been published elsewhere5 and are provided in the Supplementary Information.
Data collection
Data collected included demographic and clinical information, underlying hematologic or non-hematologic disease status, and transplantation characteristics.5
Patients with respiratory symptoms, fever and/or radiologic signs (new infiltrates on the chest X-ray or chest-computed tomography) suggestive of RC were submitted to a series of non-invasive procedures (sputum cultures, nasopharyngeal swab, urinary Ag tests and blood samples for basic biochemistry, serology and cytokine determinations). The indications to perform computed tomography and/or fiberoptic bronchoscopy have been described elsewere5 (Supplementary information).
Sample processing
Specific primers for influenza viruses types A, B and C, respiratory syncytial viruses types A and B, and adenovirus in one real time-PCR; and specific primers for parainfluenza viruses 1, 2, 3 and 4, coronaviruses 229E, and for generic detection of rhinoviruses and enteroviruses in another real time-PCR were carried out in BAL and nasopharyngeal swab when they were clinically indicated.
The methodology for obtaining culture samples, diagnostic tests for Legionella pneumophila and Streptococcus pneumoniae Ag and detection of CMV and aspergillus galactomannan Ag has been described elsewhere.5
Definitions
As explained in our previous study,5 all episodes of RC were carefully recorded and were further divided into infectious, non-infectious or undetermined. Among patients who presented more than one complication, each episode was recorded and considered independently.
Respiratory infections were diagnosed when clinical data suggested an infectious etiology, microbiological agents were isolated in any processed sample, and the clinical course and response to treatment were in accordance with an infectious etiology. One or more microbiologically documented bacteria in association with one or more viruses were considered a mixed infection. Pneumonia was diagnosed in cases with fever (⩾38 ºC), cough, dyspnea and/or purulent expectoration and new or progressive pulmonary infiltrate not explained by any other non-infectious cause.17 As noted above, microbiological agent isolation from respiratory samples was required for inclusion in the study.
Criteria for diagnosis of non-infectious RC are described in the Supplementary Information and published elsewhere.5
Neutropenia was defined as an ANC of <0.5 × 109 cells/L.
In accordance with Koya et al.,18 we further classified RI in two groups: bacterial–fungal infections and intracellular infection that included virus and tuberculosis.
Biomarker measurements
Blood samples were centrifuged, aliquoted and frozen at −80 °C for subsequent analyses. Levels of CRP, proADM and PCT were measured using plasma samples within the first 24–48 h after identification of the RC (day 0) and on days 3 and 7 thereafter. Concentrations of PCT and proADM were determined using sandwich immunoassays and time-resolved amplified cryptate emission (TRACE) technology (PCT sensitive KRYPTOR and proADM KRYPTOR; BRAHMS; Hennigsdorf, Germany). C-reactive protein was measured using a commercially available immunoturbidimetric method (Siemens Medical Solutions Diagnostics, Tarrytown, NY, USA).
The minimum detection level was 0.02 ng/mL for PCT; 0.23 nmol/L for proADM and 0.1 mg/dL for CRP. The cutoff value was 0.50 ng/mL for PCT; 0.55 nmol/L for proADM and 1 mg/dL for CRP.
Cytokine measurements
Determination of TNF-α, interleukin (IL)-1β, IL-6, IL-8 and IL-10 ultrasensitive (US) was performed in serum samples on the first day. Cytokine levels were measured by enzyme linked immunoassay (Diasource Inmunoassays, Louvain-la-Neuve, Belgium) and IL-10 US with an ultrasensitive assay (Invitrogen Coorporation, Camarillo, CA, USA). Detection limits were 0.7 pg/mL for TNF-α; 0.2 pg/mL for IL-10 US and 2 pg/mL for IL-1β, IL-6 and IL-8. The values taken as the upper limits were 10 pg/mL for TNF-α; 5 pg/mL for IL-6; 8.2 pg/mL for IL-10 US and 15 pg/mL for IL-1β and IL-8.
Statistical analysis
Continuous variables were summarized as mean±s.d. or median (interquartile range) and compared using the t-test or the non-parametric Mann–Whitney test, respectively. Categorical variables were expressed as n (%) and compared using the χ2-test or the Fisher exact test when appropriate. In order to establish the predictive value of biomarkers for the different RC, the area under the curve in the receiver operating characteristic analysis was calculated.
Univariate analyses that included clinical variables associated with an increased risk of mortality identified in our previous study5 and concentrations of the different biomarkers and cytokines studied on day 0 were performed. Variables that showed a significant result in the univariate analysis (P<0.1) were included in the initial Cox proportional hazards model.19 A manual backward stepwise selection was used to determine factors associated with 1-year mortality. The hazard ratio and 95% confidence interval (CI) were calculated. Proportional hazards assumptions were tested using log-minus-log plots. To investigate the lack of fit of our final model we evaluated the deviance residuals. The level of significance was set at 0.05 (2-tailed). All statistical analyses were done using IBM SPSS Statistics version 20.0 (Armonk, New York, USA).
Results
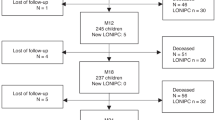
During the study period, a total of 102 episodes of RC were identified. Of these, 64 episodes corresponding to 51 patients were finally included in the study (22 episodes were not included due to undetermined or mixed etiology and 16 were not evaluated due to lack of samples; Figure 1). Patient characteristics are reported in Table 1.
Forty-five episodes of RC were of infectious origin (70%) and 19 episodes were due to non-infectious RC (30%). Most of RC occurred during the first 6 months after HSCT (87% of RI and 84% of non-infectious RC).
Viruses were the most common microorganisms causing RI (58%) (Table 2). In particular, Rhinovirus was the most commonly involved virus (35% of viral RI). Most of viral infections (73%) occurred in autumn and winter. Bacteria were the second most common agents causing RI (27%), and Pseudomonas aeruginosa was the microorganism most commonly involved (50% of the documented bacterial RI). Five of the cases with microbiologically documented bacterial infections had positive blood cultures. Finally, fungi represented 15% of RI.
Engraftment syndrome (26%), pulmonary edema (21%) and underlying disease progression (21%) were the most common non-infectious etiologies (Table 2). Twenty of the identified episodes of RC occurred in patients with neutropenia (31%), 12 in patients with acute GvHD (19%) and 10 in patients with chronic GvHD (16%). Thirty-eight (59%) and 52 (81%) samples were obtained in patients who were on immunosuppressive medication and antibiotics, respectively.
Biomarkers and cytokines in infectious and non-infectious respiratory complications
Table 3 shows biomarker and cytokine levels at different times in the population evaluated. There were no significant differences in leukocytes or neutrophil counts between infectious and non-infectious RC. A significant increase was observed in levels of CRP and proADM on day 0 in the non-infectious group (P=0.005 and P=0.03; respectively). By contrast, between-group cytokine levels were similar, except IL-6 on day 0, which was significantly higher in non-infectious complications (P=0.04).
A good correlation was observed between levels of PCT, CRP (P<0.001) and proADM (P<0.01).
Biomarker/cytokine levels did not differ between patients with and without neutropenia for both groups of complications.
Biomarkers and cytokines in respiratory infections
Levels of CRP, PCT and proADM on day 0 were higher in bacterial–fungal infections compared with viral infections (CRP, 10.46±8.49 mg/dL vs 6.52±8.15 mg/dL (P=0.02); PCT, 11.39±22 ng/mL vs 0.47±0.49 ng/mL (P=0.04); proADM, 3.62±3.30 nmol/L vs 1.65±1.03 nmol/L (P<0.01)). As shown in Figure 2, levels of CRP, PCT and proADM tended to decrease over time (days 0, 3 and 7) in bacterial–fungal RI, whereas they were persistently low in viral infections. To analyze the discriminatory power of biomarkers for the detection of bacterial–fungal RI, a receiver operating characteristic curve analysis was calculated for each biomarker (Figure 3). ProADM had the highest area under the curve (0.75, 95% CI 0.60–0.91; P<0.01). A value ⩾2.125 nmol/L of proADM on day 0 was associated with bacterial–fungal RI with a sensitivity of 61%, a specificity of 83%, a positive predictive value (PPV) of 79%, and an negative predictive value (NPV) of 68%. We also tested whether persistently low biomarkers values could rule out bacterial–fungal RI. For various cutoff levels of the maximum value obtained on days 0, 3 and 7, we calculated, for each biomarker, sensitivity, specificity, PPV and NPV for precluding systemic bacterial–fungal infection. The results show that persistently lower levels (for the first 7 days) of CRP and proADM can be very specific in ruling out bacterial–fungal infections. In particular, levels of CRP below 4.07 mg/dL had a sensitivity of 46%, a specificity of 94% and a PPV of 92% and levels of proADM below 0.91 had a sensitivity of 28%, a specificity and a PPV of 100% (Table 4). A combination of PCT (⩾1.695 ng/mL) and proADM (⩾2.245 nmol/L) confirmed a bacterial–fungal infection with a sensitivity of 28%, a specificity of 100%, a PPV of 100%, and an NPV of 58%.
Biomarker levels on days 0, 3 and 7 in bacterial–fungal respiratory infections compared with viral respiratory infections. In each box plot, the median value is indicated by the center horizontal line, and the 25th and 75th percentiles are indicated by the lower and upper box horizontal lines. Whiskers above and below the box indicate the 90th and 10th percentiles. Mann–Whitney test for comparisons of two groups.
Baseline levels of cytokines evaluated did not differ among the different RI.
Biomarkers and cytokines and clinical characteristics
Clinical variables such as type of transplantation (allogeneic vs autologous), intensity of conditioning, and GvHD did not influence biomarker/cytokine levels for either non-infectious or infectious PC (Tables 1, 2 and 3 of Supplementary Information).
Outcome
Overall 1-year mortality was 33% (17 out of 51 patients). Sixty-five per cent of all cause mortality was due to RC (11 out of 17 cases). Procalcitonin and proADM levels on day 0 were significantly higher in patients who died compared with survivors (Figure 4). In addition, CRP values showed a significant decrease over time (days 0, 3 and 7) in survivors compared with deceased patients (P=0.001).
Biomarker levels on day 0 in survivors compared with deceased patients. In each box plot, the median value is indicated by the center horizontal line, and the 25th and 75th percentiles are indicated by the lower and upper box horizontal lines. Whiskers above and below the box indicate the 90th and 10th percentiles. Mann–Whitney test for comparisons of two groups.
Univariate analysis showed that Max-PCT (+1 mg/mL, P=0.012), Max-proADM (+1 mg/mL, P=0.002), Max-CRP (+1 mg/dL, P=0.010), GvHD (P=0.019) and infection of bacterial–fungal etiology (P=0.021) were risk factors associated with 1-year mortality. Of these variables, infection of bacterial–fungal etiology (hazard ratio 6.24, 95% CI 1.32–29.47; P=0.021) was the only variable associated with 1-year mortality in the multivariate analysis.
Discussion
The results of this prospective study show that although biomarkers cannot differentiate between infectious and non-infectious complications in HSCT recipients, they still can be useful from a clinical standpoint. Levels of CRP, PCT and proADM on day 0 reflect the severity of the RC and have prognostic relevance. Moreover, persistent low levels of these biomarkers suggest a viral infection and may help to tailor the therapeutic regime.
In immunocompetent patients, biomarkers such as PCT and proADM are helpful in the diagnosis and management of severe bacterial infections.20, 21 Also, in febrile neutropenic patients, high serum PCT and proADM levels have been associated with the diagnosis and the severity of bacterial–fungal infection.22, 23 It has been speculated that in HSCT recipients, biomarkers might be of particularly utility due to the range of potential complications, both infectious and non-infectious, that these patients may suffer. Previous studies evaluating different biomarkers in this setting have been controversial.15, 24, 25, 26, 27 Pihusch et al.24 evidenced that CRP, PCT and IL-6 levels increase in both infectious and non-infectious complications. Furthermore, these authors showed that plasma levels of PCT were useful for differentiating between infection and other transplant-related complications, although, no details were given on the etiology and localization of the infection, making it difficult to draw comparisons with our own series. By contrast, a meta-analysis that included six different studies and a total of 1344 episodes of suspected systemic infection in the immediate post-transplant period demonstrated that CRP and PCT were neither sensitive nor specific enough for discriminating systemic infection from other inflammatory complications.26 However, variations in clinical criteria used to define severe systemic infection across studies and the different etiology of fever among the groups compared were an evident source of heterogeneity.28 The present study confirms the poor discriminative value of these biomarkers for diagnosing RI in this cohort of patients. In fact, the highest values of some of these biomarkers were found in patients with non-infectious RC. These findings are relevant as one of the strengths of the present study is that, contrary to other series, the episodes have been collected prospectively during a 1 year follow-up from a specific cohort of HSCT recipients. Furthermore, we have included only patients with RC and a specific etiology.
In this sense, a valid concern with the published trials involving both immunocompetent and immunosuppressed patients is the paucity of data that associate the microbiologic etiology of respiratory tract infections with biomarker levels.29
In this series, patients with non-infectious complications had significantly higher levels of CRP, proADM and IL-6 than patients with RI. A potential reason for this is that pathways that cause the secretion of biomarkers after infection can also be activated by other inflammatory conditions. Hematopoietic SCT recipients with GvHD can exhibit PCT positivity due to gastrointestinal damage causing translocation of lipopolysaccharide and secondary production of inflammatory cytokines.15, 26 Moreover, engraftment syndrome, an inflammatory condition occurring during granulocyte recovery in HSCT and characterized by fever without an identifiable infectious source and manifestations of vascular leak, is associated with elevated concentrations of several cytokines and CRP.30, 31, 32, 33 In our series, 26% of the non-infectious RC was due to the engraftment syndrome, a situation that could partially explain the high levels of biomarkers found in these patients.15
When considering only RI, our results confirm previous findings that biomarkers were significantly increased in bacterial–fungal RI compared with viral etiology.16
A possible application of biomarkers in clinical practice is their potential high NPV when measured repetitively to rule out specific infections. In this regard, our results confirm those obtained by Koya et al.18 in a small series of 28 patients, demonstrating that persistent low values of biomarkers, particularly CRP and proADM, almost certainly rule out a bacterial–fungal infection as the etiologic agent of the RI in these patients, information that may encourage the physician in charge to withhold specific antibiotic prescriptions with potential serious adverse effects.34 Other authors have found that levels of PCT allow for discrimination between episodes of fever caused by bacteria, fungi and parasites from fever of viral etiologies.35 Reasons for the absence of elevation of these biomarkers in viral infections are not well understood, but it has been shown that, by secreting interferon-gamma, viruses induce an inhibitory effect on PCT gene transcription.36 Thus, the combination of aggressive attempts at pathogen detection together with determination of biomarker levels would enhance antibiotic stewardship, particularly considering the high prevalence of viral pulmonary infections in HSCT recipients.29
Another potential utility of biomarkers in clinical management is the assessment of the severity of the complication.18, 25, 37, 38 In the present study, levels of PCT and proADM were significantly higher in patients who died during RC compared with survivors. Other authors have also found an association between levels of PCT and mortality in critically ill patients.38, 39 Koya et al.18 also identified this association in a small series of HSCT recipients. This information can be very useful when making decisions about intensive care unit admission and monitoring of the patients.
Different limitations must be considered for the proper evaluation of this study. First, the lack of a control group of HSCT recipients without RC is a flaw. Certainly, HSCT recipients undergo different stages (aplastic phase, recovery phase), and suffer some conditions (acute and/or chronic graft) or complications (presence of mucositis) that may potentially influence biomarker levels. Nevertheless, the demonstration of significant differences between the different types of complications confirm that the use of biomarkers can be useful, as a complementary tool in the management of RC in HSCT recipients. Second, we decided to apply rigorous exclusion criteria and include only those cases with microbiologically documented RI. The authors are aware of the relatively small number of patients included, which was mainly due to the numerous dropouts of the primary cohort owing to the lack of identification of the causal microorganism or, conversely, owing to the identification of a mixed infection (caused by several microorganisms). Most of the studies published on this topic include patients with both clinical and microbiological diagnosis of infection. This fact represents a potential bias, since RI may be produced by different microorganisms or by a combination of them, thus making it impossible to correlate a specific microorganism with biomarker levels.18 Third, in our study we divided, as other authors did, extracellular from intracellular infections. We are aware of the conflicting results obtained by different authors regarding the biomarkers behavior in fungal diseases. Although controversial, it is possible that biomarker levels may differ between fungal and bacterial pulmonary infections and further studies are needed on this issue. Finally, most of the patients included in this study were receiving antibiotic and/or immunosuppressive therapy when biomarker values were measured, and we cannot therefore assess the potential influence of these variables on biomarker concentrations.
In summary, in HSCT recipients, CRP, PCT and proADM cannot differentiate between infectious and non-infectious RC. However, these biomarkers can estimate the severity of infection. Persistently low values of these biomarkers militate against a bacterial–fungal infection, and an alternative diagnosis such as viral infections should be considered in the appropriate clinical setting.
References
Gratwohl A, Baldomero H, Aljurf M, Pasquini MC, Bouzas LF, Yoshimi A et al. Hematopoietic stem cell transplantation: a global perspective. JAMA 2010; 303: 1617–1624.
Ho VT, Weller E, Lee SJ, Alyea EP, Antin JH, Soiffer RJ . Prognostic factors for early severe pulmonary complications after hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2001; 7: 223–229.
Soubani AO, Pandya CM . The spectrum of noninfectious pulmonary complications following hematopoietic stem cell transplantation. Hematol Oncol Stem Cell Ther 2010; 3: 143–157.
Afessa B, Peters SG . Major complications following hematopoietic stem cell transplantation. Semin Respir Crit Care Med 2006; 27: 297–309.
Lucena CM, Torres A, Rovira M, Marcos MA, de la Bellacasa JP, Sánchez M et al. Pulmonary complications in hematopoietic SCT: a prospective study. Bone Marrow Transplant 2014; 49: 1293–1299.
Rañó A, Agustí C, Jimenez P, Angrill J, Benito N, Danés C et al. Pulmonary infiltrates in non-HIV immunocompromised patients: a diagnostic approach using non-invasive and bronchoscopic procedures. Thorax 2001; 56: 379–387.
Smith RP, Lipworth BJ, Cree IA, Spiers EM, Winter JH . A clinical marker in community-acquired pneumonia. Chest 1995; 108: 1288–1291.
Chalmers JD, Singanayagam A, Hill AT . C-reactive protein is an independent predictor of severity in community-acquired pneumonia. Am J Med 2008; 121: 219–225.
Matwivoff GN, Prahl JD, Miller RJ, Carmichael JJ, Amundson DE, Seda G et al. Immune regulation of procalcitonin: a biomarker and mediator of infection. Inflamm Res 2012; 61: 401–409.
Kutz A, Briel M, Christ-Crain M, Stolz D, Bouadma L, Wolff M et al. Prognostic value of procalcitonin in respiratory tract infections across clinical settings. Crit Care 2015; 19: 74.
España PP, Capelastegui A, Mar C, Bilbao A, Quintana JM, Diez R et al. Performance of pro-adrenomedullin for identifying adverse outcomes in community-acquired pneumonia. J Infect 2015; 70: 457–466.
Sato M, Nakasone H, Oshima K, Ishihara Y, Wada H, Sakamoto K et al. Prediction of transplant-related complications by C-reactive protein levels before hematopoietic SCT. Bone Marrow Transplant 2013; 48: 698–702.
Sjöqvist C, Snarski E . Inflammatory markers in patients after hematopoietic stem cell transplantation. Arch Immunol Ther Exp 2013; 61: 301–307.
Schots R, Kaufman L, Van Riet I, Ben Othman T, De Waele M, Van Camp B et al. Proinflammatory cytokines and their role in the development of major transplant-related complications in the early phase after allogeneic bone marrow transplantation. Leukemia 2003; 17: 1150–1156.
Mori Y, Miyawaki K, Kato K, Takenaka K, Iwasaki H, Harada N et al. Diagnostic value of serum procalcitonin an C-reactive protein for infections after allogeneic hematopoietic stem cell transplantation versus nontransplant setting. Intern Med 2011; 50: 2149–2155.
AI Shuaibi M, Bahu RR, Chaftari AM, AI Wohoush I, Shomali W, Jiang Y et al. Pro-adrenomedullin as a novel biomarker for predicting infections and response to antimicrobials in febrile patients with hematologic malignancies. Clin Infect Dis 2013; 56: 943–950.
Aguilar-Guisado M, Jiménez-Jambrina M, Espigado I, Rovira M, Martino R, Oriol A et al. Pneumonia in allogeneic stem cell transplantation recipients: a multicenter prospective study. Clin Transplant 2011; 25: E629–E638.
Koya J, Nannya Y, Ichikawa M, Kurokawa M . The clinical role of procalcitonin in hematopoietic SCT. Bone Marrow Transplant 2012; 47: 1326–1331.
Collett D . Modelling Survival Data in Medical Research. Chapman and Hall: London, UK, 1994.
Christ-Crain M, Müller B . Procalcitonin in bacterial infections—hype, hope, more or less? Swiss Med Wkly 2005; 135: 451–460.
Bello S, Lasierra AB, Mincholé E, Fandos S, Ruiz MA, Vera E et al. Prognostic power of proadrenomedullin in community-acquired pneumonia is independent of aetiology. Eur Respir J 2012; 39: 1144–1155.
Sakr Y, Sponholz C, Tuche F, Brunkhorst F, Reinhart K . The role of procalcitonin in febrile neutropenic patients: review of the literature. Infection 2008; 36: 396–407.
Giamarellos-Borboulis EJ, Grecka P, Poulakou G, Anargyrou K, Katsilambros N, Giamarellou H . Assessment of procalcitonin as a diagnostic marker of underlying infection in patients with febrile neutropenia. Clin Infect Dis 2001; 32: 1718–1725.
Pihusch M, Pihusch R, Fraunberger P, Pihusch V, Andreesen R, Kolb HJ et al. Evaluation of C-reactive protein, interleukin-6, and procalcitonin levels in allogeneic hematopoietic stem cell recipients. Eur J Haematol 2006; 76: 93–101.
Hambach L, Eder M, Dammann E, Schrauder A, Sykora KW, Dieterich C et al. Diagnostic value of procalcitonin serum levels in comparison with C-reactive protein in allogeneic stem cell transplantation. Haematologica 2002; 87: 643–651.
Blijlevens NM, Donnelly JP, Meis JF, De Keizer MH, De Pauw BE . Procalcitonin does not discriminate infection from inflammation after allogeneic bone marrow transplantation. Clin Diagn Lab Immunol 2000; 7: 889–892.
Agustí C, Rañó A, Rovira M, Filella X, Benito N, Moreno A et al. Inflammatory response associated with pulmonary complications in non-HIV immunocompromised patients. Thorax 2004; 59: 1081–1088.
Lyu YX, Yu XC, Zhu MY . Comparison of the diagnostic value of procalcitonin and C-reactive protein after hematopoietic stem cell transplantation: a systematic review and meta-analysis. Transpl Infect Dis 2013; 15: 290–299.
Gilbert DB . Where do we go from here? J Infect Dis 2015; 212: 1687–1689.
Spitzer TR . Engraftment syndrome: double-edged sword of hematopoietic cell transplants. Bone Marrow Transplant 2015; 50: 469–475.
Takatsuka H, Takemoto Y, Yamada S, Wada H, Tamura S, Fujimori Y et al. Complications after bone marrow transplantation are manifestations of systemic inflammatory response síndrome. Bone Marrow Transplant 2000; 26: 419–426.
Spitzer TR . Engraftment syndrome following hematopoietic stem cell transplantation. Bone Marrow Transplant 2001; 27: 893–898.
Chang L, Frame D, Braun T, Gatza E, Hanauer DA, Zhao S et al. Engraftment syndrome after allogeneic hematopoietic cell transplantation predicts poor outcomes. Biol Blood Marrow Transplant 2014; 20: 1407–1417.
Vincent JL, Teixeira L . Sepsis biomarkers. Value and limitations. Am J Respir Crit Care Med 2014; 190: 1081–1082.
Karzai W, Oberhoffer M, Meier-Hellmann A, Reinhart K . Procalcitonin–a new indicator of the systemic response to severe infections. Infection 1997; 25: 329–334.
Linscheid P, Seboek D, Nylen ES, Langer I, Schlatter M, Becker KL et al. In vitro and in vivo calcitonin I gene expression in parenchymal cells: a novel product of human adipose tissue. Endocrinology 2003; 144: 5578–5584.
Debiane L, Hachem RY, AI Wohoush I, Shomali W, Bahu RR, Jiang Y et al. The utility of proadrenomedullin and procalcitonin in comparison to C-reactive protein as predictors of sepsis and bloodstream infections in critically ill patients with cancer. Crit Care Med 2014; 42: 2500–2507.
Sauer M, Tiede K, Fuchs D, Gruhn B, Berger D, Zintl F . Procalcitonin, C-reactive protein, and endotoxin after bone marrow transplantation: identification of children at high risk of morbidity and mortality from sepsis. Bone Marrow Transplant 2003; 31: 1137–1142.
Habarth S, Holeckova K, Froidevaux C, Pittet D, Ricou B, Grau GE et al. Diagnostic value of procalcitonin, interleukin-6, and interleukin-8 in critically ill patients admitted with suspected sepsis. Am J Respir Crit Care Med 2001; 164: 396–402.
Acknowledgements
CML and CA contributed to the study design, the acquisition, analysis and interpretation of data, and writing of the manuscript. AT and MR participated in the study design and in the revision of the article. XF and MS contributed to the interpretation of data. RD contributed to the acquisition of data. AG conducted the statistical analysis. The project was supported by FIS-ISCIII (Fondo de Investigación Sanitaria, Instituto de Salud Carlos III) (PI041199), SEPAR 2010 and FUCAP 2010. CML was supported by a clinical research grant from SEPAR 2012 and Hospital Clínic Barcelona, Spain (Beca Josep Font 2009-2012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Bone Marrow Transplantation website
Supplementary information
Rights and permissions
About this article
Cite this article
Lucena, C., Rovira, M., Gabarrús, A. et al. The clinical value of biomarkers in respiratory complications in hematopoietic SCT. Bone Marrow Transplant 52, 415–422 (2017). https://doi.org/10.1038/bmt.2016.280
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bmt.2016.280
This article is cited by
-
Comparison of procalcitonin and C-reactive protein as early diagnostic marker for the identification of transplant-related adverse events after allogeneic hematopoietic stem cell transplantation in pediatric patients
Journal of Cancer Research and Clinical Oncology (2019)