Abstract
Background:
Tumour stroma has important roles in the development of colorectal cancer (CRC) metastasis. We aimed to clarify the roles of microRNAs (miRNAs) and their target genes in CRC stroma in the development of liver metastasis.
Methods:
Tumour stroma was isolated from formalin-fixed, paraffin-embedded tissues of primary CRCs with or without liver metastasis by laser capture microdissection, and miRNA expression was analysed using TaqMan miRNA arrays.
Results:
Hierarchical clustering classified 16 CRCs into two groups according to the existence of synchronous liver metastasis. Combinatory target prediction identified tenascin C as a predicted target of miR-198, one of the top 10 miRNAs downregulated in tumour stroma of CRCs with synchronous liver metastasis. Immunohistochemical analysis of tenascin C in 139 primary CRCs revealed that a high staining intensity was correlated with synchronous liver metastasis (P<0.001). Univariate and multivariate analyses revealed that the tenascin C staining intensity was an independent prognostic factor to predict postoperative overall survival (P=0.005; n=139) and liver metastasis-free survival (P=0.001; n=128).
Conclusions:
Alterations of miRNAs in CRC stroma appear to form a metastasis-permissive environment that can elevate tenascin C to promote liver metastasis. Tenascin C in primary CRC stroma has the potential to be a novel biomarker to predict postoperative prognosis.
Similar content being viewed by others
Main
Colorectal cancer (CRC) is the third most commonly diagnosed cancer, the burden of which has been increasing, and one of the major causes of cancer death worldwide (Ferlay et al, 2015; Arnold et al, 2017). The liver is the most common site of metastasis in CRC, developing in 40–50% of patients who have undergone resection of the primary tumour (Garden et al, 2006), and liver metastasis is still one of the major determinants of survival. To control this metastatic disease and improve patient survival, studies need to further elucidate the mechanisms underlying CRC liver metastasis.
Emerging evidence suggests important roles of the stromal components in cancer, orchestrating with each other to promote primary tumourigenesis to metastasis. Furthermore, the tumour stroma forming a heterogeneous tumour microenvironment may be a novel target for anticancer therapy (Hanahan and Weinberg, 2011; Hanahan and Coussens, 2012). Although many previous reports have demonstrated the important roles of tumour–stroma interactions in the formation of CRC metastasis, all the functions of the tumour stroma have not been revealed because it is complex and includes a variety of cellular components.
MicroRNAs (miRNAs) are small non-coding RNAs consisting of 18–25 nucleotides, which regulate the expression of genes involved in many physiological conditions, including cancer (Garzon et al, 2010). Alterations of miRNA expression in the tumour microenvironment can occur by direct interactions of stromal and cancer cells (Kohlhapp et al, 2015). A number of previous studies have conducted gene and miRNA expression analyses in cancer tissue, most of which used whole tumour tissues, mainly fresh frozen samples (Lin et al, 2011; Zhang et al, 2012). However, many studies have shown that stromal cells, including fibroblasts and inflammatory cells, are present in the vast majority (up to 90%) of tumour tissues (Dvorak, 1986), and miRNAs are differentially expressed between tumour cells and stroma in primary CRC tissue (Della Vittoria Scarpati et al, 2014). Therefore, many of the previous studies on miRNA expression in whole tumour tissue appear to have a limitation, and it is unclear how the heterogeneity of tumour tissues has affected their gene expression analysis.
Laser capture microdissection (LCM) is important to select more cancer cell-specific or stroma-specific targets that predict a poor prognosis of CRCs (Nishida et al, 2012; Calon et al, 2015). Emphasising on the value of the LCM method, we have previously analysed differences in miRNA expression between cancer cells at primary CRCs and liver metastases, and reported an important role of liver-specific miR-122 and concomitant suppression of its target gene cationic amino-acid transporter 1 (CAT1) in the primary CRC to promote liver metastasis (Iino et al, 2013). A following study comparing miRNA expression between primary CRCs and liver metastasis using LCM reported that the most significantly differentially expressed miRNA was miR-122, which was consistent with our findings (Ellermeier et al, 2014). A recent study conducted miRNA array analysis of cancer cells in T1 stage CRCs using LCM, and developed a ‘three-miRNA classifier’ to predict lymph node metastasis (Jung et al, 2016). Previous studies using LCM have consistently reported important roles of miRNAs in cancer cells in CRC development and metastasis. However, in contrast to miRNA studies of cancer cells, only few studies have focused on miRNA expression in CRC stroma (Kang et al, 2015; Pecqueux et al, 2016).
In this study, we revealed the miRNA expression profiles in tumour stroma by miRNA array analysis using LCM to identify miRNAs and their target genes as novel molecular markers involved in CRC liver metastasis.
Materials and methods
Patients and tumours
For miRNA array analysis, we used formalin-fixed, paraffin-embedded (FFPE) specimens of eight primary CRCs without metastasis, four CRCs with synchronous liver metastasis, and four CRCs with metachronous liver metastasis. (Supplementary Table S1). For immunohistochemical analysis, we used FFPE tissues of 139 primary CRCs. All samples were obtained from primary CRCs resected at Hamamatsu University Hospital (Hamamatsu, Japan) between January 2003 and December 2007. No patients had received any anticancer therapy before surgery. Ethical approval for the study was obtained from the Institutional Review Board (IRB). The IRB agreed that no written consent was required from patients for the use of FFPE tissue blocks and anonymised clinicopathological data.
Laser capture microdissection
To extract RNA from FFPE tissues, samples were microdissected using a laser microdissection system (LMD 6000; Leica Microsystems, Wetzlar, Germany) as described previously (Iino et al, 2013). The paraffin blocks were cut into 16 μm-thick sections and placed onto Leica RNase free PEN slides (Leica, Bannockburn, IL, USA). The FFPE sections were deparaffinised in xylene before hydration in a graded series of ethanol solutions and then lightly stained with 0.1% cresyl violet using an LCM staining kit (Ambion, Austin, TX, USA), followed by immediate microdissection. The tissue was collected in 50 μl digestion buffer from a mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Yokohama, Japan). A representative tissue section used for LMD is shown in Supplementary Figure S1.
RNA extraction
Total RNA was extracted from the tissue with a RecoverAll kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. The quality and quantity of total RNA were assessed with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
MicroRNA array analysis
Reverse transcription (RT) was carried out using 7.2 ng total RNA as starting material, Megaplex RT Primers, Human Pool A (Thermo Fisher Scientific) as RT primers, a TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific), and GeneAmp PCR System 9700 (Thermo Fisher Scientific) according to the manufacturer’s protocols. Before real-time PCR, miRNA was subjected to preamplification using Megaplex PreAmp Primers, Human Pool A (Thermo Fisher Scientific) as primers, a TaqMan PreAmp Master Mix (Thermo Fisher Scientific), and the GeneAmp PCR System 9700. Preamplified cDNA was then used for real-time PCR in TaqMan Universal Master Mix II, no UNG (Thermo Fisher Scientific) and applied to a TaqMan Array Human MicroRNA A Card (Thermo Fisher Scientific). The arrays were run on an ABI PRISM 7900HT Sequence Detection System (Thermo Fisher Scientific) according to the manufacturer’s protocol. Quantitative PCR data were analysed by ABI PRISM SDS 2.1 software using standard procedures (Thermo Fisher Scientific). Expression levels of individual miRNAs were determined by the ΔCt approach relative to the average Ct value of three normalisation controls (U6, RNU44, and RNU48).
MicroRNA target gene prediction
An online software programme, miRDB (http://www.mirdb.org/miRDB/), was used to predict target genes (Wong and Wang, 2015). The top 10 downregulated miRNAs were applied to the Target Mining mode of miRDB with a search filter for human genes.
Immunohistochemical analysis
To determine protein expression levels of tenascin C, immunochemical staining was carried out using 4 μm-thick sections of FFPE tissues. After deparaffinisation and rehydration, samples were blocked with 3% H2O2 for 5 min at room temperature. Antigen retrieval was carried out by incubation with Proteinase K (S302080, Dako, Tokyo, Japan) for 5 min at room temperature. The samples were then incubated with a primary mouse monoclonal antibody against human tenascin C (4F10TT, 1 : 6000 dilution, Immuno-Biological Laboratories, Fujioka, Japan) at 4 °C overnight. The sections were washed and then incubated with the secondary antibody (K500711, Dako) for 30 min at room temperature. Staining signals were developed using 3,3′-diaminobenzidine (K500711, Dako). Counterstaining was performed with haematoxylin, followed by mounting. The stained sections were scanned using an Aperio Digital Pathology Whole Slide Scanner (Leica Biosystems, Vista, CA, USA). Diagnosis was conducted in a virtual slide using an Aperio Image scope (Leica Biosystems). The area used for diagnosis was 1/3 from the deepest invasive margin, while the margin close to the normal colon tissue was excluded (Supplementary Figure S2). The calculated area for diagnosis was 23.3 mm2 per section in median and 1.9–105 mm2 per section in range. Three researchers including two clinical pathologists at Hamamatsu University Hospital with no information about the clinical data evaluated tenascin C immunoreactivity independently. The tenascin C staining intensity score was defined as 0, negative or obscure; 1, weak; 2, moderate; and 3, strong (Supplementary Figure S3). Smooth muscle and vessel wall staining intensities were considered as internal controls. Cases with high tenascin C staining in the internal control were subtracted by 1 point. Peritumoural staining was evaluated, and stained areas with an uncertain tendency were excluded. Tenascin C staining intensity scores evaluated by the three researchers were integrated for each case. When two or three researchers evaluated the same score, it was adopted as the final score. When three researchers evaluated different score, the median score was adopted. The tenascin C staining relativity was defined as the percentage of positive cells in stromal areas surrounding the cancer cells (Supplementary Figure S2).
Cell culture
SW620 human colon cancer cells (ECACC 87051203) were purchased from DS Pharma Biochemical (Osaka, Japan). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, WAKO, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere with 5% CO2. The CCD18-Co human colon fibroblast cells (CRL-1459) were purchased from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Eagle’s minimal essential medium (EMEM, Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% FBS at 37 °C in a humidified atmosphere with 5% CO2.
MicroRNA transfection
SW620 cells and CCD18-Co cells were grown to 30–50% confluence and then the medium was changed to serum-free DMEM and EMEM, respectively. AccuTarget miRNA mimic negative control, human miR-198 mimic, inhibitor negative control, or human inhibitor miR-198 (Bioneer, Daejeon, Korea) were transfected into the cells using serum-free Opti-MEM (Invitrogen, Carlsbad, CA, USA) and Lipofectamine RNAiMAX (Invitrogen) at a final concentration of 100 nM according to the manufacturer’s instructions. At 6 h after transfection, the medium was changed to DMEM or EMEM containing 10% FBS, and the cells were collected at 48 h after transfection. The nucleotide sequence of the human miR-198 mimic was 5′-GGUCCAGAGGGGAGAUAGGUUC-3′. The nucleotide sequence of the human miR-198 inhibitor was 5′-GGUCCAGAGGGGAGAUAGGUUC-3′.
Western blotting
Cells were lysed in chilled lysis buffer (50 mM Tris, pH 7.5, 300 mM sodium chloride, and 0.3% Triton-X 100) supplemented with a protease inhibitor cocktail (10 mg ml−1 each of antipain, leupeptin, pepstatin, trypsin inhibitor, and E64; and 2.5 mg ml−1 chymostatin). Protein concentrations were determined using Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA, USA). Protein extracts (18–60 μg) were electrophoresed through a polyacrylamide-SDS gradient gel (Kanto Chemical, Tokyo, Japan) and transferred onto a polyvinylidene difluoride membrane (MilliporeSigma, Darmstadt, Germany). The membrane was blocked in 3% dry milk and probed with a mouse monoclonal antibody against human tenascin C (4F10TT, 1 : 500; Immuno-Biological Laboratories, Fujioka, Japan) or mouse monoclonal anti-α-tubulin antibody (DM1A, 1 : 10 000; Sigma-Aldrich). Antibody–antigen complexes were visualised using anti-mouse IgG HRP Conjugate (W402B, 1 : 6666; Promega, Madison, WI, USA) and Clarity Max Western ECL Blotting Substrates (Bio-Rad). Primary and secondary antibodies were diluted in Can Get Signal Immunoreaction Enhancer Solution (TOYOBO, Osaka, Japan). Signal intensities were quantified using ImageJ 1.49v (National Institutes of Health, Bethesda, MD, USA) and normalised to the α-tubulin density.
Quantitative RT–PCR
Total RNA was isolated from cultured cells using ISOGEN (NIPPON GENE, Toyama, Japan) for mRNA or using a TRIzol reagent (Invitrogen) for miRNA. Isolated mRNA and miRNA was reverse transcribed to cDNA using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa Bio, Shiga, Japan) and a miScript II RT Kit (Qiagen, Hilden, Germany), respectively. The primer sequences were 5′-CTCCCAGTGACAACATCGCAATA-3′ and 5′-GGATGGCTTCCAATGACACATTTA-3′ for tenascin C mRNA; and 5′-GCACCGTCAAGGCTGAGAAC-3′ and 5′-TGGTGAAGACGCCAGTGGA-3′ for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The primers for non-coding small nuclear RNA U6 (Hs_RNU6-2_11) and miR-198 (Hs_miR-198_3) were purchased from Qiagen. The expression of mRNA and miRNA was detected using a SYBR Premix Ex Taq II (TaKaRa Bio) and a miScript SYBR Green PCR Kit (Qiagen), respectively. Quantitative RT–PCR was performed and analysed using a Thermal Cycler Dice Real Time System II Sequence detection system and software (TaKaRa Bio). Tenascin C and miR-198 transcripts were normalised to GAPDH mRNA and U6 snRNA, respectively.
Statistical analysis
The χ2-test was used for univariate analysis to evaluate the association between tenascin C expression and the existence of synchronous liver metastasis. The Kaplan–Meier method and log-rank test were used to estimate postoperative overall survival and postoperative liver metastasis-free survival. The Cox proportional hazard model was used for univariate and multivariate analyses to assess the associations of clinicopathological factors with postoperative overall survival and postoperative liver metastasis-free survival. P-values of <0.05 were considered as statistically significant. All calculations were performed with the statistical package SPSS 22.0 (SPSS Inc., Chicago, IL, USA).
Results
MicroRNA array analysis of tumour stroma classifies 16 CRCs into two groups according to the existence of synchronous liver metastasis
To elucidate the difference in miRNA expression profiles among tumour stromas of primary CRCs with or without liver metastasis, miRNA array analyses were carried out using extracts from eight CRCs without liver metastasis and four CRCs each with synchronous or metachronous liver metastasis. Patient profiles are shown in Supplementary Table S1. Tumour stroma was dissected from FFPE tissues using LCM methods. As shown in Supplementary Figure S4, hierarchical clustering classified 16 CRCs into two groups according to the existence of synchronous liver metastasis. Four CRC patients with metachronous liver metastasis and eight CRC patients without liver metastasis showed similar miRNA expression profiles in their stromal tissues, which were distinct from those in CRCs with synchronous liver metastasis (Supplementary Figure S4).
Of 377 miRNAs analysed, only 2 miRNAs, miR-29c and miR-195, were significantly upregulated in tumour stroma of CRCs with synchronous liver metastasis compared with those without synchronous liver metastasis (Table 1). In contrast, 14 miRNAs were significantly downregulated in tumour stroma of CRCs with synchronous liver metastasis. Among them, miR-302a, miR-551b, miR-627, miR-19a, miR-372, miR-384, and miR-198 were expressed at certain levels in all 12 CRCs without synchronous liver metastasis, but not in any of the 4 CRCs with synchronous liver metastasis.
MicroRNA target gene prediction identifies tenascin C as a candidate gene involved in CRC liver metastasis
MicroRNA functions as a guide by base pairing with target mRNA to negatively regulate its expression (Bartel, 2004). MicroRNAs have a role in coordinating the cancerous network by regulating many different pathways and expression of genes (Garzon et al, 2010). We assumed that some miRNAs downregulated in tumour stroma of CRCs with synchronous liver metastasis might cooperatively upregulate metastasis-promoting factors. Thus, combinatorial miRNA-targeting prediction was conducted using the Target Mining mode of miRDB (Wong and Wang, 2015). Because up to 10 miRNAs can be used for Target Mining in miRDB, the top 10 miRNAs downregulated in tumour stroma of CRCs with synchronous liver metastasis, the fold change of which was <0.03 (Table 1), were applied to the analysis. Table 2 shows the top 30 of 739 targets predicted in the analysis. Among them, we decided to focus on tenascin C, a predicted target of miR-198, as a candidate therapeutic target or novel prognostic marker that may be involved in the formation of CRC liver metastasis, because its cancer-promoting characteristics have been reported in previous studies on many malignancies (Lowy and Oskarsson, 2015; Yoshida et al, 2015). The predicted miR-198 target sequence is located at position 569–575 of the tenascin C 3′-untranslated region in human chromosome 9q33.1 (Supplementary Figure S5).
Expression levels of tenascin C protein in the primary CRC stroma correlate with the existence of synchronous liver metastasis
Expression levels of tenascin C protein were immunohistochemically analysed in 139 primary CRC samples, and the tenascin C staining intensity and relativity were evaluated as described in the Materials and Methods (Supplementary Figures S2 and S3). Clinicopathological features, including the tenascin C staining intensity and relativity, were analysed according to the existence of synchronous liver metastasis. As shown in Supplementary Table S2, lymph node metastasis, venous invasion, tumour stage, and the tenascin C staining intensity were significantly correlated with synchronous liver metastasis, while tenascin C staining relativity was comparable between CRCs with or without synchronous liver metastasis.
Expression levels of tenascin C protein in the primary CRC stroma correlate with venous invasion
We next analysed relationships between the expression levels of tenascin C protein and clinicopathological factors in 139 primary CRCs. There were positive correlations between the tenascin C staining intensity and tumour depth, lymph node metastasis, poor differentiation, venous invasion, tumour stage, and tenascin C staining relativity (Supplementary Table S3). There were also positive correlation between tenascin C staining relativity and tumour depth, lymphatic invasion, venous invasion, tumour stage, and tenascin C staining intensity (Supplementary Table S4). Among the analysed clinicopathological factors, venous invasion was strongly correlated with both the tenascin C staining intensity (P=0.001) and relativity (P<0.001; Supplementary Tables S3 and S4).
Tenascin C protein is highly expressed in tumour stroma of liver metastasis compared with primary CRC stroma
To address whether tenascin C protein is highly expressed in tumour stroma of liver metastasis, we stained tenascin C in 32 CRC liver metastases surgically resected without chemotherapy and 51 liver metastases treated with preoperative chemotherapy followed by hepatectomy. Tenascin C staining intensities in the tumour stroma of 83 CRC liver metastases were compared with those of 139 primary CRCs. As shown in Supplementary Figure S6, the tenascin C staining intensity in tumour stroma was higher in liver metastases than in primary CRCs independently of preoperative chemotherapy.
Tenascin C immunohistochemical status in primary CRC stroma correlates with poor postoperative outcomes
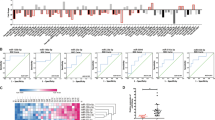
Because the staining intensity of tenascin C in CRC stroma was correlated with synchronous liver metastasis, it may be a strong postoperative prognostic marker of CRCs. To evaluate the diagnostic value of tenascin C staining in CRC stroma, we analysed the correlation of the tenascin C staining intensity and relativity with postoperative overall survival and liver metastasis-free survival of CRC patients. Of 11 CRCs with synchronous liver metastasis, 10 tumours (90.1%) showed intermediate or high tenascin C staining intensities (score 2 or 3), whereas 97 (75.8%) of 128 CRCs without synchronous liver metastasis were negative or showed a low staining intensity (score 0 or 1; Supplementary Table S2). Therefore, we classified the tenascin C staining intensities into two groups: 0–1 (n=98) and 2–3 (n= 41), and tenascin C staining relativity was also classified into two groups: <70% (n=90) and ⩾70% (n=49). CRC patients with a high tenascin C staining intensity had significantly shorter postoperative overall survival (P<0.001; Figure 1A), whereas tenascin C staining relativity was not correlated with overall survival (P=0.225; Figure 1B). Of 139 CRC patients, 128 patients did not have synchronous liver metastasis at surgery for primary CRC and their postoperative liver metastasis-free survival was analysed. Colorectal cancer patients with a high tenascin C staining intensity had significantly shorter postoperative liver metastasis-free survival (P<0.001; Figure 1C), whereas the tenascin C staining relativity was not correlated with liver metastasis-free survival (Figure 1D; P=0.159).
Survival probability of CRC patients according to tenascin C expression. (A and B) Overall survival indicated by the tenascin C staining intensity (A) and relativity (B) in tumour stroma of 139 primary CRCs. (C and D) Liver metastasis-free survival indicated by the tenascin C staining intensity (C) and relativity (D) in tumour stroma of 128 primary CRCs without synchronous liver metastasis.
Tenascin C staining intensity in primary tumour stroma is an independent prognostic marker of CRCs
We next analysed the relationships of clinicopathological factors, including the tenascin C staining intensity and relativity, with CRC patient survival to clarify prognostic factors after primary resection. Overall survival was evaluated using Cox’s proportional hazard model for univariate analysis. As shown in Table 3, the tumour depth, existence of lymph node metastasis, pathological tumour stage, postoperative chemotherapy, and tenascin C staining intensity were correlated with shorter overall survival. These factors were then applied to subsequent multivariate analysis. Multivariate analysis revealed that the tenascin C staining intensity was the only independent prognostic factor to predict postoperative overall survival (hazard ratio (HR): 3.41; 95% confidence interval (CI): 1.45–8.07; P=0.005; Table 3).
Next, postoperative liver metastasis-free survival was evaluated by univariate and multivariate analyses. Univariate analysis revealed that histopathological differentiation, postoperative chemotherapy, and the tenascin C staining intensity were correlated with shorter liver metastasis-free survival (Table 4). Multivariate analyses revealed that tenascin C staining intensity was an independent prognostic factor to predict postoperative liver metastasis-free survival (HR: 6.02; 95% CI: 2.04–17.86; P=0.001; Table 4).
Tenascin C protein expression does not correlate with its mRNA levels and can be regulated by miR-198
Although tenascin C was a predicted target of miR-198 in our analysis using the Target Mining mode of miRDB, there was no direct evidence showing that miR-198 suppresses tenascin C protein expression. To analyse whether high expression of tenascin C protein in tumour stroma is due to high levels of tenascin C mRNA, we carried out in situ hybridisation of tenascin C mRNA in 16 FFPE samples used for tenascin C immunohistochemistry (Supplementary Materials and Methods). The intensity of tenascin C mRNA was evaluated as positive or negative. Tenascin C mRNA was present at the tumour stroma in some cases and absent in others independently of tenascin C protein expression as evaluated by immunohistochemistry (Supplementary Figure S7), and there was no correlation between tenascin C mRNA expression and the staining intensity (Supplementary Table S5). Because there was no correlation between mRNA and protein expression, it was hypothesised that tenascin C might be translationally regulated perhaps in part by miRNAs, including miR-198. To address the link between miR-198 and tenascin C, we first checked expression levels of miR-198 in the 16 samples in miRNA array data, and compared them with tenascin C staining intensities. Because miR-198 was not detected in four CRC samples, while moderate miR-198 levels were consistently expressed in the remaining 12 CRC samples (average Ct value: 32.9±1.2), we classified the CRC samples according to their miR-198 expression. The CRC samples expressing miR-198 tended to have lower levels of tenascin C expression in comparison with those without miR-198 expression, although the difference was not statistically significant (P=0.237; Supplementary Figure S8). Next, to evaluate the biological link between miR-198 and tenascin C protein, we transfected a miR-198 mimic or a miR-198 inhibitor into SW620 human colon cancer cells and analysed the expression change of tenascin C protein by western blotting. As expected, relative protein expression of tenascin C was suppressed by treatment with the miR-198 mimic (Figure 2A) and increased by treatment with the miR-198 inhibitor (Figure 2B). Moreover, these changes were not observed at the mRNA level (Figure 2C). Finally, we confirmed the effect of miR-198 mimic treatment on tenascin C expression in CCD-18Co human colon fibroblast cells. These cells express much lower levels of miR-198 than SW620 cells (Figure 2D). Consistent with our observation using SW620 cells, the relative protein expression of tenascin C was suppressed by treatment with the miR-198 mimic in CCD-18Co cells (Figure 2E).
Effect of miR-198 mimic or inhibitor on tenascin C expression. (A and B) Western blotting for tenascin C and α-tubulin proteins in SW620 cells treated with a control miRNA or miR-198 mimic (A), or treated with a control or miR-198 inhibitor (B). Band densities were quantified and tenascin C protein levels were normalised to α-tubulin. Numbers at the bottom, mean fold change relative to control. (C) Quantitative RT–PCR analysis of tenascin C mRNA in SW620 cells treated with a control miRNA, miR-198 mimic, miRNA inhibitor control, or miR-198 inhibitor. Data are normalised to internal human GAPDH mRNA. All reactions were performed in triplicate and data represent the mean±s.d. for n=3 independent experiments. (D) Quantitative RT–PCR analysis of miR-198 in SW620 cells and CCD-18Co cells. Data are normalised to internal human U6 small nuclear RNA. All reactions were performed in triplicate and data represent the mean±s.d. for n=3 independent experiments. (E) Western blotting for tenascin C and α-tubulin proteins in CCD-18Co cells treated with a control miRNA or miR-198 mimic. Band densities were quantified and tenascin C protein levels were normalised to α-tubulin. Numbers at the bottom, mean fold change relative to control.
Discussion
Finding mechanisms that promote liver metastasis in CRC is important. In the era of emphasised importance of the tumour microenvironment for cancer progression, we were prompted to find specific metastasis-promoting factors in the cancer stroma. The FFPE tissues are convenient samples for retrospective research, but mRNA and protein modification caused by formalin fixation is a critical obstacle (Masuda et al, 1999). The fact that the quality of miRNA detection does not differ between FFPE and fresh frozen samples (Xi et al, 2007; Szafranska et al, 2008) led us to analyse miRNA expression using FFPE samples in the current study.
In the miRNA array analysis, we hypothesised that miRNA expression profiles in tumour stroma of CRCs with metachronous liver metastasis would be similar to those of CRCs with synchronous liver metastasis, and distinct from those without liver metastasis. However, hierarchical clustering classified 16 CRCs into two groups according to the existence of synchronous liver metastasis, and miRNA expression profiles of CRCs with metachronous liver metastasis were similar to those without liver metastasis (Supplementary Figure S4). Tumours are known to metastasise to other unrelated organs via a systemic process not only through multiple molecular events in the primary tumour cells but also by non-malignant host cells at both primary and secondary sites (Steeg, 2016). Although it is unclear whether a micrometastasis persists without complications in patients or grows progressively (Steeg, 2016), previous studies have reported the contribution of secreted molecules and bone marrow-derived endothelial progenitors to the development of macroscopic metastasis (Gao et al, 2008; Cox et al, 2016). There appears to be several models for the development of macroscopic metastasis. Tumour cells acquire additional mutations and/or associate with other tissue components and recruit bone marrow-derived cells to develop macroscopic metastasis after the formation of a microscopic metastasis, or tumour cells acquire a series of mutations in metastasis virulence genes at the primary site and directly form a macroscopic metastasis (Valastyan and Weinberg, 2011). It appears that the tumour stroma at the primary site has an important role in these processes by educating tumour cells at the primary site early in the invasion-metastasis cascade and possibly by affecting the metastatic site via the secretion of certain metastasis-promoting factors. MicroRNAs altered in the tumour stroma of CRCs with synchronous liver metastasis in the present study might affect the expression of metastasis-promoting factors to develop macroscopic liver metastasis.
In our previous study, we reported an important role of liver-specific miR-122 and concomitant suppression of its target gene CAT1 in cancer cells at the primary CRC, which promote liver metastasis (Iino et al, 2013). Increased expression of miR-122 in a metastatic liver tumour and the clinical importance of miR-122 as a prognostic marker for postoperative liver metastasis were then confirmed in reports by other groups (Vychytilova-Faltejskova et al, 2016; Maierthaler et al, 2017). Cancer cells are known to interact with tumour stroma that mainly consists of non-malignant cells, and direct communication between cancer cells and tumour stroma through secreted miRNAs has also been reported (Kohlhapp et al, 2015). There might be some crosstalk between cancer cells and tumour stroma via the stromal miRNAs identified as related to liver metastasis in the present study as well as miRNAs including miR-122 reported in previous studies on cancer cells.
Among the numerous candidates, tenascin C was selected as a suitable target for further study. In previous studies, tenascin C has been reported as a direct target of miR-335 and miR-489 in rats and mice, respectively (Chen et al, 2011; Olave et al, 2016). In the present study, neither miR-335 nor miR-489 was significantly downregulated in the tumour stroma of CRCs with synchronous liver metastasis, while tenascin C was predicted to be a target of miR-198. However, biological evidence of a relationship between miR-198 and tenascin C expression has not been reported previously. In the experiment using SW620 cells, we confirmed that protein expression of tenascin C was suppressed by treatment with a miR-198 mimic and increased by treatment with a miR-198 inhibitor. Tenascin C suppression by treatment with the miR-198 mimic was further confirmed in the experiment using CCD-18Co human colon fibroblast cells. Together, these data suggest that miR-198 suppresses tenascin C protein expression in vivo. Although further functional analysis is needed to examine whether tenascin C is a direct target of miR-198, this is the first report that demonstrates a link between miR-198 and tenascin C expression. In the in situ hybridisation experiment of tenascin C mRNA in 16 FFPE samples, tenascin C mRNA was present at the tumour stroma in some cases and absent in others independently of tenascin C protein expression (Supplementary Figure S7). It is difficult to conclude whether there is a correlation between mRNA and protein levels of tenascin C in CRC samples in general. In a previous study, tenascin C was reported to be expressed at the mRNA level in CRC stroma (Yoshida et al, 2015). Protein expression of tenascin C may not only be affected by the expression levels of tenascin C mRNA but also regulated translationally, which may involve miR-198.
Tenascin C is a glycoprotein in the extracellular matrix, which is tightly regulated in normal adult tissues (Tucker and Chiquet-Ehrismann, 2009) and expressed during embryonic development of the central nervous system and in injury sites physiologically (Chiquet-Ehrismann et al, 1986; Midwood and Orend, 2009). In cancer, it has many suggested roles in promoting metastasis, modulating adhesion and motility, facilitating angiogenesis, and establishing immune tolerance (Lowy and Oskarsson, 2015). A recent study by Li et al (2016) compared stromal proteomes at various stages of CRCs using LCM, iTRAQ labelling, and two-dimensional liquid chromatography-tandem mass spectrometry, and reported that the expression levels of S100A9 and tenascin C were correlated with TNM stages and metastasis. Because the expression levels of tenascin C protein in tumour stroma of primary CRCs were strongly correlated with venous invasion compared with other clinicopathological factors (Supplementary Tables S3 and S4), tenascin C appears to have major roles in the formation of CRC liver metastasis by upregulating invasive phenotype of cancer cells. Tenascin C has been reported to have diverse cellular roles in promoting metastasis, and the best studied and characterised functions are adhesive and counter-adhesive activities that regulate cell motility (Midwood and Orend, 2009). Previous studies demonstrated in vitro functional roles of tenascin C secretion from the stromal cells to facilitate adhesion and migration of ovarian carcinoma cells (Wilson et al, 1999), and to induce the invasion of pancreatic adenocarcinoma cells through its receptor annexin A2 (Foley et al, 2017). In CRCs, it has been reported that tenascin C produced by myofibroblasts can stimulate colon cancer cells to an invasive phenotype via RhoA inactivation (De Wever et al, 2004). Although detailed mechanisms were not fully understood, tumour–stroma interaction through tenascin C appears to induce invasive phenotype of CRCs to promote liver metastasis. In the present study, we not only confirmed the increased levels of tenascin C in CRCs with metastasis but also clarified the prognostic value of tenascin C to predict postoperative overall survival and liver metastasis-free survival. In addition, this is the first report identifying that stromal miRNAs at primary CRCs upregulate tenascin C to develop synchronous liver metastasis. Although antitumour effects need to be evaluated in future studies, the miRNA identified in the present study and tenascin C are expected to be novel therapeutic targets in CRCs.
The tumour stroma mainly consists of non-malignant cells in the tumour tissue, such as fibroblasts, immune cells, and vasculature with endothelial cells and pericytes, as well as basement membrane and extracellular matrix (Tarin, 2013; de Visser et al, 2006). In tumours, activated fibroblasts are called cancer-associated fibroblasts (CAFs) and have been reported to facilitate communication between the tumour cells and the tumour microenvironment (Franco et al, 2010). Immune cells, particularly T lymphocytes, serve as regulatory factors in the tumour microenvironment, and generalised tumour inflammatory infiltrate is a candidate marker for the prognosis of patients with CRC (Liotta and Kohn, 2001; Mei et al, 2014). Tumour vasculature has important roles in the tumour microenvironment, and blood vessels provide a niche that maintains cancer stemness (Takakura, 2012). In various tumours, CAFs in the reactive stroma are thought to be the main source of tenascin C production, and their heterogeneous distribution in cancer tissue tends to be in the invasive front (Xing et al, 2010; Lowy and Oskarsson, 2015). In immunohistochemical diagnosis concerning tenascin C, we found staining heterogeneity in tumours. Application of whole slide imaging using virtual microscopy has been reported to provide a simple and precise overview of a tumour, leading to more reliable diagnosis of heterogeneous histopathology (Halama et al, 2010). Using this whole slide imaging with virtual microscopy, we found that tenascin C was mainly expressed in the peritumoural extracellular matrix at the invasive front. Although further evaluation is needed in a future study, the vast majority of stromal components producing tenascin C may be CAFs at the invasive front.
In the analysis of tenascin C and the clinicopathological characteristics, the tenascin C staining intensity was statistically correlated with the existence of synchronous liver metastasis (Supplementary Table S2). Moreover, the Cox’s proportional hazard model for multivariate analysis revealed that the tenascin C staining intensity was an independent prognostic factor to predict postoperative overall survival (Table 3) and liver metastasis-free survival (Table 4). These data are consistent with the previous finding that stromal tenascin C expression accelerates the metastatic potential (Lowy and Oskarsson, 2015). In this study, we also applied the tenascin C staining relativity to evaluate its importance in predicting CRC patient survival, because its utility has been reported in a previous study (Sis et al, 2004). Unexpectedly, the tenascin C staining relativity was not a significant prognostic factor for either overall survival or liver metastasis-free survival in our study. These findings suggest that the tenascin C staining intensity, rather than relativity, could be a reliable prognostic factor to predict postoperative overall survival and liver metastasis-free survival of CRC patients. However, intensity measurements are typically more subjective and more sensitive to immunohistochemical conditions than relative expression levels. The tenascin C staining intensity correlated with liver metastasis-free survival with a higher HR and lower P-value compared with overall survival (HR: 6.02 vs 3.41; P-value: 0.001 vs 0.005; Tables 3 and 4). Tenascin C expression in tumour stroma appears to be predominantly related to liver metastasis-specific outcomes rather than overall survival.
Although our data appear to be robust, there are some points that require further clarification. First, the number of CRC patients with synchronous liver metastasis is relatively small because most patients have received chemotherapy before surgery in recent years. Thus, our samples are largely restricted to FFPE specimens from a relatively long time ago. Second, the evaluation of immunohistochemical staining may not perfectly reflect the differential expression of miRNAs because LCM was not applied at the invasion front. Accumulating evidence suggests that tumour stromal heterogeneity may influence the variation of altered miRNAs. Therefore, a detailed study applying next-generation sequencing of miRNAs or total RNA from various areas in the tumour stroma, including the invasion front, could provide interesting and convincing findings.
In conclusion, our data add evidence supporting that heterogeneity in CRC stroma, which promotes liver metastasis, can be identified by investigating the differential expression of miRNAs. Tenascin C expression in tumour stroma was identified as a novel molecular marker of CRC to predict postoperative overall survival and liver metastasis-free survival.
Change history
26 October 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut 66 (4): 683–691.
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2): 281–297.
Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, Sevillano M, Palomo-Ponce S, Tauriello DV, Byrom D, Cortina C, Morral C, Barcelo C, Tosi S, Riera A, Attolini CS, Rossell D, Sancho E, Batlle E (2015) Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 47 (4): 320–329.
Chen C, Wu CQ, Zhang ZQ, Yao DK, Zhu L (2011) Loss of expression of miR-335 is implicated in hepatic stellate cell migration and activation. Exp Cell Res 317 (12): 1714–1725.
Chiquet-Ehrismann R, Mackie EJ, Pearson CA, Sakakura T (1986) Tenascin: an extracellular matrix protein involved in tissue interactions during fetal development and oncogenesis. Cell 47 (1): 131–139.
Cox TR, Gartland A, Erler JT (2016) Lysyl oxidase, a targetable secreted molecule involved in cancer metastasis. Cancer Res 76 (2): 188–192.
de Visser KE, Eichten A, Coussens LM (2006) Paradoxical roles of the immune system during cancer development. Nat Rev Cancer 6 (1): 24–37.
De Wever O, Nguyen QD, Van Hoorde L, Bracke M, Bruyneel E, Gespach C, Mareel M (2004) Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac. FASEB J 18 (9): 1016–1018.
Della Vittoria Scarpati G, Calura E, Di Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De Stefano A, Pepe S, De Placido S, D'Incalci M, Marchini S, Carlomagno C (2014) Analysis of differential miRNA expression in primary tumor and stroma of colorectal cancer patients. BioMed Res Int 2014: 840921.
Dvorak HF (1986) Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med 315 (26): 1650–1659.
Ellermeier C, Vang S, Cleveland K, Durand W, Resnick MB, Brodsky AS (2014) Prognostic microRNA expression signature from examination of colorectal primary and metastatic tumors. Anticancer Res 34 (8): 3957–3967.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136 (5): E359–E386.
Foley K, Muth S, Jaffee E, Zheng L (2017) Hedgehog signaling stimulates tenascin C to promote invasion of pancreatic ductal adenocarcinoma cells through Annexin A2. Cell Adh Migr; e-pub ahead of print 2 February 2017; doi:10.1080/19336918.2016.1259057.
Franco OE, Shaw AK, Strand DW, Hayward SW (2010) Cancer associated fibroblasts in cancer pathogenesis. Semin Cell Dev Biol 21 (1): 33–39.
Gao D, Nolan DJ, Mellick AS, Bambino K, McDonnell K, Mittal V (2008) Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science 319 (5860): 195–198.
Garden OJ, Rees M, Poston GJ, Mirza D, Saunders M, Ledermann J, Primrose JN, Parks RW (2006) Guidelines for resection of colorectal cancer liver metastases. Gut 55 (Suppl 3): iii1–iii8.
Garzon R, Marcucci G, Croce CM (2010) Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 9 (10): 775–789.
Halama N, Zoernig I, Spille A, Michel S, Kloor M, Grauling-Halama S, Westphal K, Schirmacher P, Jager D, Grabe N (2010) Quantification of prognostic immune cell markers in colorectal cancer using whole slide imaging tumor maps. Anal Quant Cytol Histol 32 (6): 333–340.
Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21 (3): 309–322.
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144 (5): 646–674.
Iino I, Kikuchi H, Miyazaki S, Hiramatsu Y, Ohta M, Kamiya K, Kusama Y, Baba S, Setou M, Konno H (2013) Effect of miR-122 and its target gene cationic amino acid transporter 1 on colorectal liver metastasis. Cancer Sci 104 (5): 624–630.
Jung CK, Jung SH, Yim SH, Jung JH, Choi HJ, Kang WK, Park SW, Oh ST, Kim JG, Lee SH, Chung YJ (2016) Predictive microRNAs for lymph node metastasis in endoscopically resectable submucosal colorectal cancer. Oncotarget 7 (22): 32902–32915.
Kang WK, Lee JK, Oh ST, Lee SH, Jung CK (2015) Stromal expression of miR-21 in T3-4a colorectal cancer is an independent predictor of early tumor relapse. BMC Gastroenterol 15: 2.
Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME (2015) MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 34 (48): 5857–5868.
Li M, Peng F, Li G, Fu Y, Huang Y, Chen Z, Chen Y (2016) Proteomic analysis of stromal proteins in different stages of colorectal cancer establishes Tenascin-C as a stromal biomarker for colorectal cancer metastasis. Oncotarget 7 (24): 37226–37237.
Lin M, Chen W, Huang J, Gao H, Ye Y, Song Z, Shen X (2011) MicroRNA expression profiles in human colorectal cancers with liver metastases. Oncol Rep 25 (3): 739–747.
Liotta LA, Kohn EC (2001) The microenvironment of the tumour-host interface. Nature 411 (6835): 375–379.
Lowy CM, Oskarsson T (2015) Tenascin C in metastasis: a view from the invasive front. Cell Adh Migr 9 (1–2): 112–124.
Maierthaler M, Benner A, Hoffmeister M, Surowy H, Jansen L, Knebel P, Chang-Claude J, Brenner H, Burwinkel B (2017) Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer 140 (1): 176–187.
Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K (1999) Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 27 (22): 4436–4443.
Mei Z, Liu Y, Liu C, Cui A, Liang Z, Wang G, Peng H, Cui L, Li C (2014) Tumour-infiltrating inflammation and prognosis in colorectal cancer: systematic review and meta-analysis. Br J Cancer 110 (6): 1595–1605.
Midwood KS, Orend G (2009) The role of tenascin-C in tissue injury and tumorigenesis. J Cell Commun Signal 3 (3-4): 287–310.
Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Sugihara K, Doki Y, Mori M (2012) Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res 18 (11): 3054–3070.
Olave N, Lal CV, Halloran B, Pandit K, Cuna AC, Faye-Petersen OM, Kelly DR, Nicola T, Benos PV, Kaminski N, Ambalavanan N (2016) Regulation of alveolar septation by microRNA-489. Am J Physiol Lung Cell Mol Physiol 310 (5): L476–L487.
Pecqueux M, Liebetrau I, Werft W, Dienemann H, Muley T, Pfannschmidt J, Mussle B, Rahbari N, Scholch S, Buchler MW, Weitz J, Reissfelder C, Kahlert C (2016) A comprehensive microRNA expression profile of liver and lung metastases of colorectal cancer with their corresponding host tissue and its prognostic impact on survival. Int J Mol Sci 17: 10.
Sis B, Sagol O, Kupelioglu A, Sokmen S, Terzi C, Fuzun M, Ozer E, Bishop P (2004) Prognostic significance of matrix metalloproteinase-2, cathepsin D, and tenascin-C expression in colorectal carcinoma. Pathol Res Pract 200 (5): 379–387.
Steeg PS (2016) Targeting metastasis. Nat Rev Cancer 16 (4): 201–218.
Szafranska AE, Davison TS, Shingara J, Doleshal M, Riggenbach JA, Morrison CD, Jewell S, Labourier E (2008) Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling. J Mol Diagn 10 (5): 415–423.
Takakura N (2012) Formation and regulation of the cancer stem cell niche. Cancer Sci 103 (7): 1177–1181.
Tarin D (2013) Role of the host stroma in cancer and its therapeutic significance. Cancer Metastasis Rev 32 (3-4): 553–566.
Tucker RP, Chiquet-Ehrismann R (2009) The regulation of tenascin expression by tissue microenvironments. Biochim Biophys Acta 5: 888–892.
Valastyan S, Weinberg RA (2011) Tumor metastasis: molecular insights and evolving paradigms. Cell 147 (2): 275–292.
Vychytilova-Faltejskova P, Pesta M, Radova L, Liska V, Daum O, Kala Z, Svoboda M, Kiss I, Slaby O (2016) Genome-wide microRNA expression profiling in primary tumors and matched liver metastasis of patients with colorectal cancer. Cancer Genomics Proteomics 13 (4): 311–316.
Wilson KE, Bartlett JM, Miller EP, Smyth JF, Mullen P, Miller WR, Langdon SP (1999) Regulation and function of the extracellular matrix protein tenascin-C in ovarian cancer cell lines. Br J Cancer 80 (5–6): 685–692.
Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43 (Database issue): 5.
Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J (2007) Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 13 (10): 1668–1674.
Xing F, Saidou J, Watabe K (2010) Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci (Landmark Ed) 15: 166–179.
Yoshida T, Akatsuka T, Imanaka-Yoshida K (2015) Tenascin-C and integrins in cancer. Cell Adh Migr 9 (1-2): 96–104.
Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P, Zhang Q, Dong L, Liu Y, Dong J (2012) microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep 27 (3): 685–694.
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research in Japan 15K10132 (H Kikuchi) and 15H04929 (H Konno). We thank Drs Satoshi Sakai, Masaki Baba, Yuki Kurita, and Yayoi Kawabata for their advice.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Murakami, T., Kikuchi, H., Ishimatsu, H. et al. Tenascin C in colorectal cancer stroma is a predictive marker for liver metastasis and is a potent target of miR-198 as identified by microRNA analysis. Br J Cancer 117, 1360–1370 (2017). https://doi.org/10.1038/bjc.2017.291
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.291
Keywords
This article is cited by
-
Spatial profiling of cancer-associated fibroblasts of sporadic early onset colon cancer microenvironment
npj Precision Oncology (2023)
-
Transposon-activated POU5F1B promotes colorectal cancer growth and metastasis
Nature Communications (2022)
-
Desmoplastic reactions and epithelial-mesenchymal transition proteins in stages II and III colorectal cancer: association with and prognostic value for disease-free survival
Virchows Archiv (2022)
-
Tenascin-C expression in the lymph node pre-metastatic niche in muscle-invasive bladder cancer
British Journal of Cancer (2021)
-
Differential regulation of extracellular matrix proteins in three recurrent liver metastases of a single patient with colorectal cancer
Clinical & Experimental Metastasis (2020)