Abstract
Background:
Tumour aggressiveness might be related to the degree of main cancer hallmark acquirement of tumour cells, reflected by expression levels of specific biomarkers. We investigated the expression of Aldh1, Survivin, and EpCAM, together reflecting main cancer hallmarks, in relation to clinical outcome of colorectal cancer (CRC) patients.
Methods:
Immunohistochemistry was performed using a tumour tissue microarray of TNM (Tumour, Node, Metastasis)-stage I–IV CRC tissues. Single-marker expression or their combination was assessed for associations with the clinical outcome of CRC patients (N=309).
Results:
Increased expression of Aldh1 or Survivin, or decreased expression of EpCAM was each associated with poor clinical outcome, and was therefore identified as clinically unfavourable expression. Analyses of the combination of all three markers showed worse clinical outcome, specifically in colon cancer patients, with an increasing number of markers showing unfavourable expression. Hazard ratios ranged up to 8.3 for overall survival (P<0.001), 36.6 for disease-specific survival (P<0.001), and 27.1 for distant recurrence-free survival (P<0.001).
Conclusions:
Our data identified combined expression levels of Aldh1, Survivin, and EpCAM as strong independent prognostic factors, with high hazard ratios, for survival and tumour recurrence in colon cancer patients, and therefore reflect tumour aggressiveness.
Similar content being viewed by others
Main
To date, treatment allocation of CRC patients is based on the TNM (Tumour, Node, Metastasis) classification system of the American Joint Committee on Cancer. Under the current classification, patient groups still show large differences in clinical outcome (Benson et al, 2004; Gunderson et al, 2010; Quirke et al, 2010). Biomarkers that can better predict patient survival and the development of recurrent disease or metastasis are therefore warranted. These biomarkers can be used to further refine the TNM-staging system to identify CRC patients who may benefit from adjuvant therapy and/or close follow-up in addition to surgery. This will contribute to an approach of personalised treatment, based on individual tumour characteristics.
Advances have been made towards the discovery of biomarkers in order to improve tumour staging. The American Society of Clinical Oncology’s Tumor Markers Expert Panel (ASCO TMEP-2006) and the European Group on Tumor Markers (EGTM) reviewed the literature on a collection of biomarkers, of which most lacked the significant and discriminative value required for clinical implementation (Duffy et al, 2003; Locker et al, 2006; Duffy et al, 2007, 2013). Combining biomarkers based on tumour biology, thereby better reflecting tumour aggressiveness, might increase their clinical discriminative and prognostic value synergistically. The cancer hallmarks, as described by Hanahan and Weinberg (Hanahan and Weinberg, 2000, 2011), are those features that a tumour needs to attain to be able to sustain itself as tumour. Tumour aggressiveness may be related to the extent to which these different cancer hallmarks are acquired throughout the tumour, reflected by expression levels of specific individual biomarkers. In this study, we investigated the expression of aldehyde dehydrogenase 1 family member A1 (ALDH1A1 or Aldh1), baculoviral IAP repeat-containing protein 5 (BIRC5 or Survivin) and epithelial cell adhesion molecule (EPCAM or EpCAM). Together the expression levels of these markers represent the degree of acquirement of the main cancer hallmarks (Hanahan and Weinberg, 2000) in CRC (Table 1). Aldh1 is a cytoplasmatic enzyme responsible for the oxidation of intracellular aldehydes (Yoshida et al, 1992; Molotkov and Duester, 2003), thereby conferring therapeutic resistance to alkylating agents (von Eitzen et al, 1994). Research suggests that Aldh1 has a role in early differentiation of stem cells and their proliferation (Chute et al, 2006) and metastasis (Huang et al, 2009). Expression of Aldh1 is associated with cancer stem cells (Corti et al, 2006). Survivin plays an important role in the regulation of apoptosis (Ambrosini et al, 1997; Kawasaki et al, 1998, 2001; Rodel et al, 2002; Williams et al, 2003; Xiaoyuan et al, 2010) and cell division (Altieri, 1994). Survivin is required for normal fetal development, and is generally no longer expressed in most adult tissues (Ambrosini et al, 1997). Re-expression of Survivin is observed in a range of human cancers (Ambrosini et al, 1997) and linked to (colorectal) carcinogenesis (Lin et al, 2003). Expression of Survivin is associated with metastasis (Rodel et al, 2002; Lassmann et al, 2007; Chu et al, 2012), local recurrent disease (Rodel et al, 2005) and poor prognosis (Kawasaki et al, 1998; Sarela et al, 2000; Rodel et al, 2002; Sprenger et al, 2011) in CRC. Membrane glycoprotein EpCAM is expressed on the highly proliferative cells of the intestinal epithelium (Balzar et al, 1999), and is overexpressed in most human carcinomas (Went et al, 2004), including colorectal carcinomas (Herlyn et al, 1979; Spizzo et al, 2011). Contradictory roles for EpCAM in cancer development have been published and reviewed (van der Gun et al, 2010). In CRC, loss of membranous EpCAM expression is generally associated with a tumour-promoting role and poor patient survival (Litvinov et al, 1994a, 1994b; Basak et al, 1998; Went et al, 2006; Gosens et al, 2007; Lugli et al, 2010).
Expression levels of specific biomarkers were hypothesised to represent the extent to which certain cancer hallmarks are acquired in individual colorectal tumours, and to correlate with clinical outcome. Based on the properties of each of the three studied biomarkers described in Table 1, above-median expression of Aldh1, above-median expression of Survivin, and below-median expression of EpCAM were hypothesised to denote clinically unfavourable (associated with poor clinical outcome) biomarker expression in colorectal tumours. In combined analyses, an increase in the number of biomarkers with unfavourable expression could imply more aggressive tumours.
In summary, we investigated tumour expression of Aldh1, survivin, and EpCAM in correlation with patient survival in order to predict clinical outcome in CRC patients.
Materials and methods
Study cohort
The study population consisted of 309 CRC patients and is described in Figure 1. Information of covariate data was available for the patients, and included age at operation, gender, TNM-stage, tumour location, tumour diameter, microsatellite stability status (MSS-status), history of cancer, adjuvant treatment, tumour recurrence, and the occurrence of a new primary tumour in the follow-up period. The follow-up period was right-censored in October 2011 or ended earlier due to death or loss to follow-up. Informed consent was obtained from all patients included in the study and the use of these specimens was approved by the Medical Ethical Committee of the Leiden University Medical Center (LUMC). All samples were coded, according to national ethical guidelines (‘Code for Proper Secondary Use of Human Tissue’, Dutch Federation of Medical Scientific Societies). This study was performed according to the REMARK guidelines (NCI-EORTC) (McShane et al, 2005).
Tissue microarrays (TMAs)
The preparation of the TMAs was described by Zeestraten et al (2014). In brief, tumour tissues were collected from a consecutive series of CRC patients who underwent surgery at the LUMC of their primary colon or rectum tumour between 1991 and 2001 (N=470) and of whom tumour tissue was available (N=409). Formalin-fixed paraffin-embedded (FFPE) samples from each of the patients were used to construct a TMA. Haematoxylin and eosin-stained tissue sections of each of the tumour tissue blocks were reviewed by a pathologist for histopathologically representative tumour regions. Per patient, three tumour tissue cores of size 0.6 mm were transferred to a recipient paraffin block using a custom-made precision instrument. Tissue sections of 4 μm were cut for immunohistochemistry.
Determination of MSS-status
Determination of MSS-status was published previously for the colon cancer patients of the patient population by Zeestraten et al (2014). This section reports the determination of the total patient cohort, including the previously reported colon cancer patients and the rectum cancer patients. In brief, tumour tissue cores of size 2 mm were collected for all patients of whom additional FFPE material was available (N=329). Paraffin was dissolved in xylene. Tissues were rehydrated in ethanol (100 and 70%) and subsequently dried for 10 min at 37 °C. DNA was extracted using the Nucleospin 96 Tissue kit (Machery-Nagel, Düren, Germany) according to the manufacturer’s protocol. As described previously (Zeestraten et al, 2012), MSS-status was assessed using the MSI Analysis System Version 1.2 (Promega, Mannheim, Germany) and interpreted by an experienced pathologist. For 59 patients of our study cohort (N=309) MSS-status was unknown due to non-informative results or absence of additional FFPE material.
Assessment of marker expression
Antibodies against Aldh1 (ALDH1A1, AB52492, Abcam, Cambridge, UK), Survivin (BIRC5, AB469, Abcam) and EpCAM (Ab323A3, in-house produced hybridoma, kindly provided by the LUMC Department of Pathology (Edwards et al, 1986)) were used for immunohistochemistry to detect expression in the tumour cells at predetermined optimal dilutions. TMA sections of 4 μm were deparaffinised in xylene and rehydrated in a series of graded alcohol-to-(distilled)water dilutions. Antigen retrieval was performed by trypsin treatment or by heat induction at 95 °C using PT Link (Dako, Glostrup, Denmark) with a low-pH Envision FLEX target retrieval solution (pH 6.0, citrate buffer, Dako). Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide in water for 20 min. The primary antibodies were incubated overnight, followed by incubation with secondary antibodies (Envision-HRP labeled polymer anti-rabbit/anti-mouse, Dako) for 30 min. Chromogen DAB (3,3-diaminobenzidine, Dako) was used for visualisation. Subsequently, sections were counterstained with haematoxylin, dehydrated, and covered.
Stained TMA slides were scanned and analysed on the Ariol system (Leica Microsystems, Wetzlar, Germany). The percentage of tumour cells positive for cytoplasmatic Aldh1, cytoplasmatic Survivin or membranous EpCAM was determined. Percentages of tumour cells expressing cytoplasmatic Aldh1 were scored semi-automated by the Ariol system according to the manufacturer’s recommendations. Two independent (blinded) observers assessed percentages of tumour cells expressing cytoplasmatic Survivin or membranous EpCAM, with the second observer assessing at least one-third of the tumour cores. Presence of the marker was classified as the percentage of stained tumour cells with 10% increments, and including 5% and 95%. The inter-observer variability was analysed using Cohen’s kappa coefficient. A kappa >0.6 was considered as sufficient inter-observer agreement, indicating the reliability of the data. The mean percentage of positive cells of the three cores per patient (from the first observer) was used for survival analysis.
Statistical analysis
All data were analysed using the statistical package SPSS 20.0 for Windows (SPSS Inc, Chicago, IL, USA). Based on the skewed distributions of percentages of positive cells, the median percentages were used as cutoff values to divide patients into two groups for each of the individual markers (single-marker analyses). Multivariate single-marker analyses were used to verify hypothesised clinically unfavourable single-marker expression. For combined-marker analyses, patients were divided into three groups, based on expression of single markers, according to the following grouping: patients with only clinically favourable marker expression (group 1), patients with unfavourable marker expression of one or two of the three markers (group 2), and patients with only clinically unfavourable marker expression (group 3). Group 1 served as the reference group in all combined-marker survival analyses.
Survival analyses were performed for single-marker expression and for the combination of the three markers. Overall survival (OS) was defined as the time from surgery until death by any cause. Disease-specific survival (DSS) was defined as the time from surgery until death by CRC. Distant recurrence-free survival (DRFS) was defined as the time from surgery until the diagnosis of a distant recurrence or death by cancer.
The relationship between single-marker expression or combined-marker expression and established prognostic factors was investigated using the Pearson χ2 test. The Cox proportional hazards model was used to analyse the association between single markers or their combination and patient survival. For OS, Kaplan–Meier curves were used to visualise these associations. For DSS and DRFS, cumulative incidence curves were calculated, accounting for death due to other causes (Putter et al, 2007). All important non-subjective covariates for CRC, as described in the section on the study cohort, were included in multivariate analyses irrespective of statistical significance, to correct for potential differences and survival influence of covariate distribution between the analysis groups. Both the occurrence of a secondary CRC or other type of primary tumour in the follow-up and adjuvant treatment in the follow-up were entered as time-dependent covariates. Differences in clinical outcome between patient groups are presented as hazard ratios (HRs). All tests were two-tailed and P-values<0.05 were considered statistically significant.
Results
Marker expression in study cohort
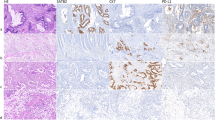
Immunohistochemical data of single-marker expression were available for over 97% of the 309 patients in the study population. The median percentages of tumour cells positive for Aldh1, Survivin and EpCAM were ∼1.6%, ∼86.5%, and ∼88.5%, respectively, which were used for subsequent group division. Figure 2 shows representative staining for below-median and above-median expression of single markers. The Cohen’s kappa coefficient of Survivin (0.66, P<0.001) and EpCAM (0.74, P<0.001) was determined as level of inter-observer agreement, showing sufficient agreement.
Examples of single-marker expression. Representative immunohistochemical staining for below-median and above-median expression (indicated below each picture) of cytoplasmatic Aldh1, cytoplasmatic Survivin, and membranous EpCAM are shown. Identical tumour cores were used for each row. In single-marker analyses, above-median expression of Aldh1 or Survivin, or below-median expression of EpCAM was identified as unfavourable in terms of clinical outcome in CRC. Representative staining of clinically favourable (A) and unfavourable (B) marker expression are indicated.
Correlation of individual marker expression with standard clinicopathological parameters is described in Table 2. Aldh1 expression was significantly correlated with age at operation, with the above-median expression group containing less elderly patients. Expression of Survivin was significantly correlated with TNM-stage, with the above-median expression group containing more patients with higher TNM-stages. Expression of EpCAM showed significant correlation with the occurrence of a primary tumour in the follow-up period, with the above-median expression group containing more patients with new primary tumours. Correlation of combined-marker expression with standard clinicopathological parameters is described in Supplementary File S1. Combined-marker expression did not show any significant correlation with standard patient or tumour characteristics.
Impact of single-marker expression on patient survival
Results of univariate and multivariate analyses are shown in Supplementary File S2. Based on the properties of Aldh1, Survivin and EpCAM presented in Table 1, above-median expression of Aldh1, above-median expression of Survivin, and below-median expression of EpCAM were hypothesised to have clinically unfavourable biomarker expression and to be associated with poor clinical outcome in colorectal tumours. Data from the multivariate single-marker analyses were used to verify clinically more favourable vs less favourable marker expression (Figure 3). Univariate analyses showed a significant association between the above-median expression of Aldh1 and poor clinical outcome in DRFS, and a trend towards a similar association for OS and DSS was identified. A significant association between above-median Survivin expression and poor clinical outcome was shown in univariate analyses for DSS and DRFS, and a trend for OS towards a similar association was identified. No significant association was found between EpCAM expression and clinical outcome in univariate analyses. In multivariate analyses, Aldh1 expression was identified as an independent prognostic factor for OS, DSS, and DRFS. For Survivin, a significant association was observed for DSS and DRFS and a trend towards an association for OS. In contrast to univariate analyses, multivariate analyses identified EpCAM expression as an independent prognostic factor for DSS. A trend towards an association between below-median EpCAM expression and poor survival was identified for OS. Together, these data show that above-median expression of Aldh1 or Survivin, or below-median expression of EpCAM, was associated with poor survival and higher tumour recurrence rates, recognising these expression patterns as clinically unfavourable phenotypes, according to expectations.
Multivariate single-marker expression analyses. Shown are the hazard ratios (HR; vertical axis; log2 scale) resulting from the different single-marker multivariate survival analyses indicated with ▪, and 95% confidence intervals indicated by protruding black lines. Data are shown for all patients in the study cohort, and for patients with colon tumours or rectum tumours separately. HR>1 indicates worse clinical outcome for above-median expression, HR<1 indicates worse clinical outcome for below-median expression. Abbreviations: A=all patients; C=patients with colon carcinoma; DRFS=distant recurrence-free survival; DSS=disease-specific survival; OS=overall survival; R=patients with rectum carcinoma.
Associations of single-marker expression with clinical outcome in multivariate analyses are illustrated in Figure 3 for the whole CRC patient cohort and for colon and rectal cancer patients separately. For Aldh1 and Survivin expression, we observed a statistical difference in clinical outcome between patients with a primary colon or rectum tumour (Supplementary File S2). For colon tumours (N=232), both uni- and multivariate analyses of OS, DSS, and DRFS showed a significant association or trend towards such an association between above-median biomarker expression and worse clinical outcome. This statistical difference was not observed in patients with rectum tumours (N=73). No difference in survival between patients with colon and rectum tumours was observed for EpCAM expression. The differences between colon and rectum tumours do not seem attributable to MSS-status, as the combined-marker expression was prognostic for colon cancer patients with microsatellite stable as well as microsatellite instable tumours (Supplementary File S3).
Impact of combined-marker expression on CRC patient survival
Combination of biomarkers, based on tumour biology, may better reflect tumour aggressiveness and might increase the clinical discriminative and prognostic value. More aggressive tumours are likely to display clinical unfavourable expression of a higher number of biomarkers. Therefore, expression of Aldh1, Survivin, and EpCAM was combined into three patient groups (as described in the Materials and Methods section) and correlated to clinical outcome. We hypothesised that a higher group number, with a higher number of clinically unfavourable marker expression, correlated with poorer clinical outcome. As expected, combined-marker expression was associated with worse clinical outcome in CRC patients (Table 3).
Because of the observed difference in survival between patients with colon or rectum tumours in single-marker analyses, we investigated this possibility for the combination of markers as well. Indeed, for colon tumours the combination of markers proved to be a very strong prognostic factor, whereas for rectum tumours the combination of markers had no prognostic value (Figure 4 and Table 3). Associations of combined-marker expression with patient survival and tumour recurrence are illustrated with Kaplan–Meier curves or cumulative incidence curves (Figure 5). Univariate survival analyses showed a significant association for OS, DSS, and DRFS between combined-marker expression and clinical outcome in colon cancer patients. A higher group number, with an increasing number of markers showing clinically unfavourable marker expression, was associated with worse survival and higher distant recurrence rates. Combined-marker expression remained an independent prognostic factor for clinical outcome for colon cancer patients in multivariate analyses of OS, DSS, and DRFS. The high HRs (ranging up to 36.6) emphasise the prognostic value of combining biomarkers for the prediction of clinical outcome (Table 3).
Multivariate combined-marker expression analyses. Shown are the hazard ratios (vertical axis; log2 scale) resulting from multivariate survival analyses of combined-marker expression indicated with ▪, and 95% confidence intervals indicated by protruding black lines. Numbers at the x-axis represent the combined-marker analysis groups, with all favourable expression in group 1, unfavourable expression of 1 or 2 markers in group 2, and all unfavourable expression in group 3. Data are shown for patients with colon tumours (N=232) or rectum tumours (N=73) separately. Abbreviations: OS=overall survival; DSS=disease-specific survival; DRFS=distant recurrence-free survival.
Curves for univariate survival analyses of combined-marker expression. Kaplan–Meier curves or cumulative incidence plots illustrating univariate survival analyses of combined-marker expression. Data are shown for patients with colon tumours (N=232) and rectum tumours (N=73) separately. Tables below the curves indicate the numbers at risk per group for the different time points. P-values of the univariate Cox proportional hazard analyses are presented in the graphs. Patients were divided in 3 groups, with all favourable expression in group 1 ( ), unfavourable expression of one or two markers in group 2 (
), unfavourable expression of one or two markers in group 2 ( ), and all unfavourable expression in group 3 (
), and all unfavourable expression in group 3 ( ).The x-axis represents survival in years since surgery. Abbreviations: DRFS=distant recurrence-free survival; DSS=disease-specific survival; OS=overall survival.
).The x-axis represents survival in years since surgery. Abbreviations: DRFS=distant recurrence-free survival; DSS=disease-specific survival; OS=overall survival.
Thus, in combined biomarker analyses, an increasing number of the biomarkers with clinically unfavourable expression in colon tumours indicated worse clinical outcome.
Discussion
In this study, we demonstrated that the combination of tumour expression levels of Aldh1, Survivin, and EpCAM is a strong predictor, with high HRs, for distant tumour recurrence and shorter survival in colon cancer patients. Our analyses implicated more aggressive tumours when an increasing number of biomarkers, representing main cancer hallmarks, show clinically unfavourable expression.
Many different processes are involved in carcinogenesis, involving many key proteins. The hallmarks of cancer, as described by Hanahan and Weinberg (Hanahan and Weinberg, 2000), represent properties that a cell needs to attain in order to become and sustain itself as a tumour cell. We hypothesised that tumour aggressiveness is related to the degree of expression of each cancer hallmark by a tumour. The strength of our biomarker collective is that we did not focus on a single process, but chose and combined markers representative of main cancer hallmarks (Hanahan and Weinberg, 2000) in colon cancer. Combining biomarkers that represent the major cancer hallmarks was based on the idea that the underlying tumour biology, and thereby tumour characteristics, could identify patients with an aggressive tumour phenotype, who may benefit from adjuvant therapy and/or close follow-up in addition to standard treatment based on the current TNM-staging guidelines. Our study is the first to report the prognostic value of this focused combination analyses of biomarkers, in a cohort of TNM-stage I–IV colon cancer patients. Our analyses shows that a higher degree of expression of the main cancer hallmarks in colon cancer cells is indeed associated with more aggressive tumours.
High expression of Aldh1 was expected to be associated with a worse clinical outcome in colon cancer patients based on its function. We are the first to show this association in a colon cancer patient cohort in multivariate survival analysis, as other studies did not reach statistical significance mostly due to low patients numbers (Lugli et al, 2010; Kahlert et al, 2012). According to our expectations, high expression of Survivin was related to poor clinical outcome in our colon cancer patient cohort. This was in line with results reported for OS in a smaller set of CRC patients (Xiaoyuan et al, 2010). Interestingly, we demonstrated that membranous Survivin expression is only prognostic in colon cancer patients, but not in rectal cancer patients. This is in contrast to Sprenger et al (2011), who demonstrated the prognostic value of Survivin expression for DFS in rectal cancer patients. These contrast findings might be due to differences in the patient cohorts. The Sprenger cohort investigated a specific patient group, namely those who had received pre-operative radiochemotherapy. In our analyses, these patients were intentionally excluded as tumour characteristics could be changed after pre-operative therapy. Although in univariate single-marker analyses EpCAM failed to reach statistical significance in our study cohort, multivariate results indicate that reduced EpCAM expression was, as hypothesised, associated with a worse DSS and higher recurrence rates in CRC. Other studies support our findings for EpCAM (Went et al, 2006; Gosens et al, 2007; Lugli et al, 2010).
There is great controversy on colon and rectum tumours being different disease entities. In many statistical analyses colon and rectum tumours are pooled and referred to as CRC. However, there are differences in tumours arising from colon and rectum tissues (Birkenkamp-Demtroder et al, 2005; Komuro et al, 2005). The main genetic difference reported is the occurrence of microsatellite instability. In our analyses, combined-marker expression showed high significance and prognostic value in colon tumours, but not in rectum tumours, which was not attributable to MSS-status. This suggests that the representation of the different cancer hallmarks by Aldh1, Survivin, and EpCAM expression is colon cancer tissue-specific. For rectum tumours, a different set of representative biomarkers needs to be identified. This difference emphasises the increasing evidence that suggests that colon and rectum tumours could be considered different disease entities (Birkenkamp-Demtroder et al, 2005; Komuro et al, 2005).
There is debate about leaving patients with TNM-stage IV out of survival analyses, as these patients often receive treatment with palliative intent instead of curable intent. In our analyses, the prognostic value of the combined-marker expression was not restricted to TNM-stages I–III. It was remarkable that the combined-marker expression also showed prognostic value in TNM-stage IV patients. TNM-stage IV patients with only favourable expression levels of Aldh1, Survivin, and EpCAM might be those patients who may actually benefit from adjuvant therapy with curable intent. We therefore also included stage IV patients in survival analyses. In addition, patients with TNM-stage III tumours and only favourable expression of the markers might not need adjuvant treatment, whereas patients with TNM-stage I or II tumours that show unfavourable marker expression only might benefit from close follow-up and adjuvant treatment, respectively.
Previously other biomarkers have been identified with prognostic and predictive properties in colon cancer. Single markers include CEA for prognosis in especially stage II and postoperative surveillance, MSS-status for prognosis in especially stage II, KRAS for predicting response/resistance to anti-EGFR antibodies (all reviewed in Duffy et al, 2007, 2013; Kelley et al, 2011), as well as BRAF for prognosis in MSS patients (Kelley et al, 2011) and P53 and TS for prognosis in stage II/III and response to therapy (reviewed in Locker et al, 2006; Duffy et al, 2007). Additionally, two gene expression signatures are available as commercial platforms: one platform for prognosis of relapse-free survival in stage II colon cancer (Coloprint (Salazar et al, 2011; Maak et al, 2013)), and one with prognostic and predictive value in stage II and III colon cancer patients (Oncotype Dx (O'Connell et al, 2010; Gray et al, 2011)). As discussed in the introduction, most of these biomarkers lacked a significant or discriminative value required for clinical implementation or need additional prospective assessment (Duffy et al, 2003; Locker et al, 2006; Duffy et al, 2007, 2013). In contrast to most of these biomarkers, the prognostic value of the proposed combination of three biomarkers is significantly discriminative, and not limited to a particular subgroup of colon cancer patients. As immunohistochemistry is a standard method used in pathology, clinical implementation of the proposed is biomarker combination is relatively easy.
In conclusion, we showed that the combination of Aldh1, Survivin, and EpCAM expression level was a strong independent risk factor for higher distant recurrence rates and shorter survival in colon cancer patients. The proposed biomarker combination showed discriminative value combined with biological and clinical significance, with HRs up to 36.6, and should be further investigated for use in clinical setting.
Change history
10 June 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Altieri DC (1994) Splicing of effector cell protease receptor-1 mRNA is modulated by an unusual retained intron. Biochemistry 33 (46): 13848–13855.
Ambrosini G, Adida C, Altieri DC (1997) A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 3 (8): 917–921.
Balzar M, Prins FA, Bakker HA, Fleuren GJ, Warnaar SO, Litvinov SV (1999) The structural analysis of adhesions mediated by Ep-CAM. Exp Cell Res 246 (1): 108–121.
Basak S, Speicher D, Eck S, Wunner W, Maul G, Simmons MS, Herlyn D (1998) Colorectal carcinoma invasion inhibition by CO17-1A/GA733 antigen and its murine homologue. J Natl Cancer Inst 90 (9): 691–697.
Benson AB III, Schrag D, Somerfield MR, Cohen AM, Figueredo AT, Flynn PJ, Krzyzanowska MK, Maroun J, McAllister P, Van CE, Brouwers M, Charette M, Haller DG (2004) American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol 22 (16): 3408–3419.
Birkenkamp-Demtroder K, Olesen SH, Sorensen FB, Laurberg S, Laiho P, Aaltonen LA, Orntoft TF (2005) Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut 54 (3): 374–384.
Chu XY, Chen LB, Wang JH, Su QS, Yang JR, Lin Y, Xue LJ, Liu XB, Mo XB (2012) Overexpression of survivin is correlated with increased invasion and metastasis of colorectal cancer. J Surg Oncol 105 (6): 520–528.
Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, McDonnell DP (2006) Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci USA 103 (31): 11707–11712.
Corti S, Locatelli F, Papadimitriou D, Donadoni C, Salani S, Del BR, Strazzer S, Bresolin N, Comi GP (2006) Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells 24 (4): 975–985.
Duffy MJ, Lamerz R, Haglund C, Nicolini A, Kalousova M, Holubec L, Sturgeon C (2013) Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer 134 (11): 2513–2522.
Duffy MJ, van DA, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C, Topolcan O (2007) Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer 43 (9): 1348–1360.
Duffy MJ, van DA, Haglund C, Hansson L, Klapdor R, Lamerz R, Nilsson O, Sturgeon C, Topolcan O (2003) Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. Eur J Cancer 39 (6): 718–727.
Edwards DP, Grzyb KT, Dressler LG, Mansel RE, Zava DT, Sledge GW Jr, McGuire WL (1986) Monoclonal antibody identification and characterization of a Mr 43,000 membrane glycoprotein associated with human breast cancer. Cancer Res 46 (3): 1306–1317.
Gosens MJ, van Kempen LC, van de Velde CJ, van Krieken JH, Nagtegaal ID (2007) Loss of membranous Ep-CAM in budding colorectal carcinoma cells. Mod Pathol 20 (2): 221–232.
Gray RG, Quirke P, Handley K, Lopatin M, Magill L, Baehner FL, Beaumont C, Clark-Langone KM, Yoshizawa CN, Lee M, Watson D, Shak S, Kerr DJ (2011) Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol 29 (35): 4611–4619.
Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK (2010) Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol 28 (2): 264–271.
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100 (1): 57–70.
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144 (5): 646–674.
Herlyn M, Steplewski Z, Herlyn D, Koprowski H (1979) Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc Natl Acad Sci USA 76 (3): 1438–1442.
Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM (2009) Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 69 (8): 3382–3389.
Kahlert C, Gaitzsch E, Steinert G, Mogler C, Herpel E, Hoffmeister M, Jansen L, Benner A, Brenner H, Chang-Claude J, Rahbari N, Schmidt T, Klupp F, Grabe N, Lahrmann B, Koch M, Halama N, Buchler M, Weitz J (2012) Expression analysis of aldehyde dehydrogenase 1A1 (ALDH1A1) in colon and rectal cancer in association with prognosis and response to chemotherapy. Ann Surg Oncol 19 (13): 4193–4201.
Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N (1998) Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res 58 (22): 5071–5074.
Kawasaki H, Toyoda M, Shinohara H, Okuda J, Watanabe I, Yamamoto T, Tanaka K, Tenjo T, Tanigawa N (2001) Expression of survivin correlates with apoptosis, proliferation, and angiogenesis during human colorectal tumourigenesis. Cancer 91 (11): 2026–2032.
Kelley RK, Van Bebber SL, Phillips KA, Venook AP (2011) Personalized medicine and oncology practice guidelines: a case study of contemporary biomarkers in colorectal cancer. J Natl Compr Canc Netw 9 (1): 13–25.
Komuro K, Tada M, Tamoto E, Kawakami A, Matsunaga A, Teramoto K, Shindoh G, Takada M, Murakawa K, Kanai M, Kobayashi N, Fujiwara Y, Nishimura N, Hamada J, Ishizu A, Ikeda H, Kondo S, Katoh H, Moriuchi T, Yoshiki T (2005) Right- and left-sided colorectal cancers display distinct expression profiles and the anatomical stratification allows a high accuracy prediction of lymph node metastasis. J Surg Res 124 (2): 216–224.
Lassmann S, Tang L, Capanu M, Brabletz T, Schopflin A, Zur HA, Gonen M, Kemeny N, Shia J, Klimstra D, Werner M (2007) Predictive molecular markers for colorectal cancer patients with resected liver metastasis and adjuvant chemotherapy. Gastroenterology 133 (6): 1831–1839.
Lin LJ, Zheng CQ, Jin Y, Ma Y, Jiang WG, Ma T (2003) Expression of survivin protein in human colorectal carcinogenesis. World J Gastroenterol 9 (5): 974–977.
Litvinov SV, Bakker HA, Gourevitch MM, Velders MP, Warnaar SO (1994a) Evidence for a role of the epithelial glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell Adhes Commun 2 (5): 417–428.
Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO (1994b) Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol 125 (2): 437–446.
Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr (2006) ASCO 2006 update of recommendations for the use of tumour markers in gastrointestinal cancer. J Clin Oncol 24 (33): 5313–5327.
Lugli A, Iezzi G, Hostettler I, Muraro MG, Mele V, Tornillo L, Carafa V, Spagnoli G, Terracciano L, Zlobec I (2010) Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br J Cancer 103 (3): 382–390.
Maak M, Simon I, Nitsche U, Roepman P, Snel M, Glas AM, Schuster T, Keller G, Zeestraten E, Goossens I, Janssen KP, Friess H, Rosenberg R (2013) Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann Surg 257 (6): 1053–1058.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2005) Reporting recommendations for tumour marker prognostic studies (REMARK). Br J Cancer 93 (4): 387–391.
Molotkov A, Duester G (2003) Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem 278 (38): 36085–36090.
O'Connell MJ, Lavery I, Yothers G, Paik S, Clark-Langone KM, Lopatin M, Watson D, Baehner FL, Shak S, Baker J, Cowens JW, Wolmark N (2010) Relationship between tumour gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol 28 (25): 3937–3944.
Putter H, Fiocco M, Geskus RB (2007) Tutorial in biostatistics: competing risks and multi-state models. Stat Med 26 (11): 2389–2430.
Quirke P, Cuvelier C, Ensari A, Glimelius B, Laurberg S, Ortiz H, Piard F, Punt CJ, Glenthoj A, Pennickx F, Seymour M, Valentini V, Williams G, Nagtegaal ID (2010) Evidence-based medicine: the time has come to set standards for staging. J Pathol 221 (4): 357–360.
Rodel F, Hoffmann J, Distel L, Herrmann M, Noisternig T, Papadopoulos T, Sauer R, Rodel C (2005) Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res 65 (11): 4881–4887.
Rodel F, Hoffmann J, Grabenbauer GG, Papadopoulos T, Weiss C, Gunther K, Schick C, Sauer R, Rodel C (2002) High survivin expression is associated with reduced apoptosis in rectal cancer and may predict disease-free survival after preoperative radiochemotherapy and surgical resection. Strahlenther Onkol 178 (8): 426–435.
Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga J, Bruin S, Kerr D, Kuppen P, van d V, Morreau H, Van VL, Glas AM, Van't Veer LJ, Tollenaar R (2011) Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol 29 (1): 17–24.
Sarela AI, Macadam RC, Farmery SM, Markham AF, Guillou PJ (2000) Expression of the antiapoptosis gene, survivin, predicts death from recurrent colorectal carcinoma. Gut 46 (5): 645–650.
Spizzo G, Fong D, Wurm M, Ensinger C, Obrist P, Hofer C, Mazzoleni G, Gastl G, Went P (2011) EpCAM expression in primary tumour tissues and metastases: an immunohistochemical analysis. J Clin Pathol 64 (5): 415–420.
Sprenger T, Rodel F, Beissbarth T, Conradi LC, Rothe H, Homayounfar K, Wolff HA, Ghadimi BM, Yildirim M, Becker H, Rodel C, Liersch T (2011) Failure of downregulation of survivin following neoadjuvant radiochemotherapy in rectal cancer is associated with distant metastases and shortened survival. Clin Cancer Res 17 (6): 1623–1631.
van der Gun BT, Melchers LJ, Ruiters MH, de Leij LF, McLaughlin PM, Rots MG (2010) EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis 31 (11): 1913–1921.
von Eitzen U, Meier-Tackmann D, Agarwal DP, Goedde HW (1994) Detoxification of cyclophosphamide by human aldehyde dehydrogenase isozymes. Cancer Lett 76 (1): 45–49.
Went P, Vasei M, Bubendorf L, Terracciano L, Tornillo L, Riede U, Kononen J, Simon R, Sauter G, Baeuerle PA (2006) Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer 94 (1): 128–135.
Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S (2004) Frequent EpCam protein expression in human carcinomas. Hum Pathol 35 (1): 122–128.
Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, Fleming J, Tavana D, Frenkel E, Becerra C (2003) Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res 9 (3): 931–946.
Xiaoyuan C, Longbang C, Jinghua W, Xiaoxiang G, Huaicheng G, Qun Z, Haizhu S (2010) Survivin: a potential prognostic marker and chemoradiotherapeutic target for colorectal cancer. Ir J Med Sci 179 (3): 327–335.
Yoshida A, Hsu LC, Dave V (1992) Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme 46 (4–5): 239–244.
Zeestraten EC, Maak M, Shibayama M, Schuster T, Nitsche U, Matsushima T, Nakayama S, Gohda K, Friess H, van de Velde CJ, Ishihara H, Rosenberg R, Kuppen PJ, Janssen KP (2012) Specific activity of cyclin-dependent kinase I is a new potential predictor of tumour recurrence in stage II colon cancer. Br J Cancer 106 (1): 133–140.
Zeestraten EC, Reimers MS, Saadatmand S, Dekker JW, Liefers GJ, van den Elsen PJ, van de Velde CJ, Kuppen PJ (2014) Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br J Cancer 110 (2): 459–468.
Acknowledgements
We thank NG Dekker-Ensink for technical support. This study was performed within the framework of CTMM, the Center for Translational Molecular Medicine; DeCoDe project (grant 03O-101).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Goossens-Beumer, I., Zeestraten, E., Benard, A. et al. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br J Cancer 110, 2935–2944 (2014). https://doi.org/10.1038/bjc.2014.226
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.226
Keywords
This article is cited by
-
Cloning, Expression, and Purification of the Human Synthetic Survivin Protein in Escherichia coli Using Response Surface Methodology (RSM)
Molecular Biotechnology (2023)
-
Tumor-suppressive role of Smad ubiquitination regulatory factor 2 in patients with colorectal cancer
Scientific Reports (2022)
-
The Vitamin D status is associated with serum C-reactive protein and adhesion molecules in patients with renal cell carcinoma
Scientific Reports (2019)
-
Prognostic value of low CDX2 expression in colorectal cancers with a high stromal content – a short report
Cellular Oncology (2019)
-
The Prognostic Value of Cancer Stem Cell Markers (Notch1, ALDH1, and CD44) in Primary Colorectal Carcinoma
Journal of Gastrointestinal Cancer (2019)