Abstract
Background:
The expression and function of ribosomal s6 protein kinase 4 (RSK4) in renal cell carcinoma (RCC) are unknown.
Methods:
Immunohistochemistry was used to detect the expression of RSK4 in RCC, and the relationship between RSK4 expression and clinicopathological features as well as prognosis of RCC patients was statistically analysed. Ectopic RSK4 expression in RCC cell lines was performed to determine its effect on cell cycle regulation, tumour invasiveness, and metastatic capability.
Results:
RSK4 was overexpressed in RCCs (P=0.003), compared with normal tissues, and the expression varied in different RCC subtypes (P=0.021), especially in two subtypes of papillary RCCs (P=0.001). RSK4 expression was positively correlated with high pT stage (P<0.001), high Fuhrman grade (P<0.001), lymph node involvement (P<0.001), and presence of distant metastasis (P=0.039), and could predict poor outcome in RCC patients. Molecular studies showed that overexpression of RSK4 could promote cell cycle progression and enhance the invasive and metastatic capability of RCC cell lines and vice versa.
Conclusion:
The expression pattern and molecular mechanisms of RSK4 in RCCs indicate that it could be a potential independent prognostic factor and serve as a new potential therapeutic target for RCC patients.
Similar content being viewed by others
Main
Renal cell carcinoma (RCC) is the most common type of kidney cancer in adults, responsible for approximately 90% of cases (Ljungberg et al, 2011). RCC consists of various histologic subtypes, with remarkably different morphologies, immunophenotypes, genetic changes, and clinical outcomes. Most patients with RCC have a high risk of relapse and metastasis (Athar and Gentile, 2008). Moreover, RCC is resistant to conventional chemotherapy and irradiation, leading to a poor prognosis. Despite improved diagnostic techniques, it is still difficult to diagnose RCC and predict prognosis. Thus, there is a strong necessity for more sensitive diagnostic and predictive parameters.
Ribosomal S6 protein kinase 4 (RSK4), is first identified as an X-linked gene in patients with mental retardation and belongs to the RSK family, playing a role in cell growth and proliferation. The expression of RSK4 mRNA is most abundant in fetal and adult kidney and brain (Yntema et al, 1999). However, there have been few and conflicting reports describing the expression and function of RSK4 protein in human tissues. It is reported that RSK4 is a predominantly cytosolic protein with very low expression, but is constitutively activated with low basal ERK activity (Dummler et al, 2005), and RSK4 participates in p53-induced growth arrest via p21 (Berns et al, 2004). Moreover, Lopez-Vicente et al has found that RSK4 could regulate replicative and stress-induced senescence, and this senescence is reversed when RSK4 is inhibited (Lopez-Vicente et al, 2009; Lopez-Vicente et al, 2011). These findings suggest that RSK4 be a tumour-suppressor gene, which is further confirmed by the downregulation of RSK4 mRNA in colon and renal carcinomas (ME et al, 2006). It is also found that RSK4 is frequently hypermethylated in endometrial cancer cell lines and in primary endometrial cancers compared with normal endometrial tissue (Dewdney et al, 2011). On the contrary, Thakur et al (2005, 2007) have found that the expression of RSK4 mRNA is higher in transgenic mouse mammary tumours and human breast cancer tissues compared with normal mammary tissues. However, further study shows that RSK4 expression could limit the oncogenic, invasive, and metastatic potential of breast cancer cells (Thakur et al, 2008). Nevertheless, Bender and Ullrich (2012) have found that RSK4 overexpression is associated with sunitinib resistance in RCC cell lines, indicating that RSK4 may regulate treatment resistance and could promote tumour progression. Whether RSK4 promotes or inhibits tumour formation is not clear, and the expression and function of RSK4 in RCCs remain uncertain. In this study, we used tissue microarray (TMA) to study the expression of RSK4 protein in human normal and tumour tissues by immunohistochemistry, and found that RSK4 protein was weakly expressed in human normal kidney and much more strongly expressed in clear cell RCCs (ccRCCs). This indicates that RSK4 may have an oncogenic role in the development of RCC. To test this hypothesis, we evaluated the expression of RSK4 in RCC tissues, analysed the relationship between RSK4 expression and the clinicopathological features of RCC patients, and explored the potential molecular mechanisms of RSK4 in RCC cell lines.
Materials and Methods
TMA
Multiple human normal and tumour organ tissue arrays were purchased from US Biomax Inc. (FDA807-1 and FDA807-2, Rockville, MD, USA). The FDA807-1 array contained 24 types of normal human organs (3 cases for each type), including cerebrum, cerebellum, adrenal gland, ovary, pancreas, thyroid gland, hypophysis, testis, parathyroid gland, breast, spleen, tonsil, thymus, bone marrow, lung, cardiac muscle, oesophagus, stomach, small intestine, colon, liver, salivary gland, kidney, and prostate. The FDA807-2 array contained 54 tumour tissues (one case for each tumour) and 6 types (3 cases for each type) of adjacent normal organs (Supplementary Table 1).
Patients and specimens
A total of 101 consecutive patients who underwent radical nephrectomy for RCC between 2000 and 2008 were identified from the pathology archives of Xijing Hospital. The patients’ charts were reviewed and those who had pre-operative radiation or chemotherapy, or were immunocompromised, were excluded. The 101 RCC patients consisted of 75 males and 26 females with a mean age of 55 years (range: 6–79 years). The median follow-up time was 45 months (range: 1–116 months). Patients were staged using the AJCC TNM system (Edge and Compton, 2010). All tumours were graded according to the Fuhrman system (Fuhrman et al, 1982). Histological classifications of RCC were assessed based on the World Health Organization (WHO) Classification of the renal tumours (Lopez-Beltran et al, 2006), including 47 cases of ccRCC, 42 papillary RCC (PRCC), 6 chromophobe RCC (CRCC), 3 collecting duct carcinoma (CDC), and 3 medullary renal carcinoma. Normal kidney tissues were obtained from 20 hydronephrosis patients. All the slides were re-examined by two pathologists to ensure correct diagnosis. The specimen collection and study procedures were approved by the Ethics Committee of Xijing Hospital.
Immunohistochemistry
Paraffin-embedded sections as well as the TMA of 4 μm thickness were deparaffinised and treated with 3% (v/v) hydrogen peroxide to block endogenous peroxidase activity. Proteolytic antigen retrieval was performed in pepsin for 30 min at 37 °C, followed by incubation in 10% (v/v) bovine serum albumin (Sigma, St. Louis, MO, USA) in phosphate-buffered saline (PBS) at room temperature for 10 min to block the non-specific antibody-binding sites. The sections were then incubated overnight at 4 °C with the mouse monoclonal antibody against the human RSK4 proteins (1 : 75 dilution; JS-31, Santa Cruz Biotechnology, Dallas, TX, USA). Later on, a standard rapid EnVision technique (Dako, Glostrup, Denmark) was used to detect the protein conjugates and develop the colour. Finally, the sections were visualised after counterstaining with haematoxylin. Serial sections of RCC were run in parallel with the primary antibody replaced by PBS and mouse IgG1 (Santa Cruz Biotechnology) as blank and negative controls.
Evaluation of immunohistochemical staining
The sections were photographed under optical microscope (BX51, Olympus, Tokyo, Japan) and the photos were captured by the software DP2-BSW (Olympus). Immunohistochemical staining was evaluated simultaneously by three observers who had no knowledge of clinicopathological features of the patients. A consensus was reached according to Shimizu’s method (Shimizu et al, 1990). Both the distribution (the percentage of positive cells) and the intensity of staining were assessed in a semi-quantitative manner. The following system was used to score the distribution of positive cells: none (not stained)=0, focal (less than one-third of cells stained)=1, multifocal (less than two-thirds of cells stained)=2, and diffuse (most cells stained)=3. The intensity of staining was graded as follows: none (not stained)=0, mild (between 0 and 2)=1, and strong (clearly identified by × 40 magnification)=2. The scores for distribution and intensity were added and graded as follows: 0–2=(−, negative), 3–5=(+, positive).
Fresh tissues, cell lines, plasmids, and transfection
We collected 10 fresh ccRCCs and the adjacent normal tissues for the determination of RSK4 expression. The human RCC cell lines GRC-1, ACHN, and 786-O were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in RPMI 1640 (Hyclone, Thermo, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 5% CO2, at 37 °C. The pcDNA3.1/Neo-RSK4 plasmid was used to stably transfect the RCC cell lines to overexpress human RSK4. Transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. Briefly, cells were cultured in six-well plates to 90% confluence. Plasmid DNA (4.0 μg per well) was mixed with 10 μl Lipofectamine 2000 to transfect the RCC cells. After 48 h, cells were trypsinised and replated onto 10-cm culture dishes in the presence of 300 μg ml−1 G418 (Gibco). Single-cell clones were isolated for clone expansion. Each cell clone was screened using reverse transcription-PCR and western blot analysis to determine RSK4 expression at both mRNA and protein levels.
Lentivirus-based shRNA transduction
RSK4 shRNA was kindly provided by Professor Cajal (Department of Pathology, Vall d’Hebron University Hospital, Barcelona, Spain). Lentivirus-based transduction was carried out using a packaging cell line, HEK293T, for co-transfection of psPAX2 and pMD2.G (kindly donated by Professor Jian Zhang, Department of Biochemistry and Molecular Biology, Fourth Military Medical University, Xi’an, China). After two consecutive viral infections, transfected cells were selected using puromycin (1 μg ml−1, Merck, Darmstadt, Germany).
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from transfected cells and fresh tissues using Trizol (Invitrogen), according to the manufacturer’s instruction, and stored at −80 °C. RSK4 mRNA in stably transfected clones was identified using reverse transcription-PCR and the SYBR Green II kit (Takara, Shiga-ken, Japan). GAPDH gene was used as an internal control. RSK4 primers used were 5′-TGAGTGGTGGAAACTGGGACAATA-3′(F) and 5′-TGGCATGGACTGTGGTCATGAGTC -3′(R), and for GAPDH were 5′-GCACCGTCAAGGCTGAGAAC-3′(F) and 5′-TGGTGAA GACGCCAGTGGA-3′(R). The annealing temperature used was 60 °C.
Western blot
Fresh tissues were ground in liquid nitrogen for immunoblotting. Both the tissues and cells were lysed in buffer (50 mmol l−1 Tris-HCl, 150 nmol l−1 NaCl, 1 mmol l−1 EDTA, 1 mmol l−1 DTT, 0.1% Tween-20, 1 mmol l−1 phenylmethylsulfonyl fluoride, 10 mmol l−1 h-glycerophosphate, 1 mmol l−1 NaF, 2 mmol l−1 Na3VO4, 1–5 μg ml−1 leupeptin, and 1–5 μg ml−1 aprotinin). Protein was quantitated using a BCA protein assay (Pierce, Thermo). Protein aliquots (50 μg per lane) were separated using 10% SDS–PAGE, transferred onto a PVDF membrane (Millipore, Billerica, MA, USA), and visualised using chemiluminescence (Pierce, Thermo). Mouse monoclonal and goat polyclonal anti-human RSK4 (JS-31 and C-20, Santa Cruz Biotechnology) and rabbit anti-human p53, p21, phosphorylated Rb (pRb), ERK1/2, phospho-ERK1/2, matrix metalloproteinase 9 (MMP-9), and CD44 (Cell Signaling Technology, Boston, MA, USA) were used for immunoblotting.
Cell cycle analysis
The cell cycle was analysed using flow cytometry (ELITE ESP, Beckman-Coulter, Brea, CA, USA). The cells were trypsinised and centrifuged for 5 min at 1000 r.p.m. The pellets were resuspended in cold PBS and kept on ice for 5 min before analysis. NP-40 and propidium iodide (Sigma) were added to a final concentration of 0.1% and 50 mg ml−1, respectively. DNA content was measured in the Fl2 channel of a fluorescence-activated cell-sorting station using CellQuest software (BD, Franklin, NJ, USA).
Matrigel invasion assay
Cells were suspended at 5 × 105 cells per ml in 1640 medium containing 1% FBS. 100 μl of the cell suspension was added to the upper well of Transwell inserts coated with 1 mg ml−1 Matrigel (Corning, Corning, NY, USA). Six hundred μl of 1640 medium with 10% FBS was added to the lower wells. The plates were incubated for 24 h at 37 °C in 5% CO2. After incubation, the inserts were carefully lifted and cells from the upper surface were gently scraped off. The remaining cells at the bottom of the filter were fixed and stained with 0.1% crystal violet. Each experiment was repeated at least three times to ensure reproducibility of the results.
ERK pathway inhibition by small molecular inhibitors
The ERK1/2 inhibitor U0126 was purchased from CST (Boston, MA, USA) and the RSK4 inhibitor BI-D1870 was obtained from Axon Medchem, (Groningen, The Netherlands). The cells were incubated in serum-free medium for 12 h, exposed to 10 μ M U0126 or 10 μ M BI-D1870 for 60 min, and then incubated in 10% FBS for 10 min. Cells were then placed in a Matrigel invasion assay. Lysates were analysed using immunoblot analysis with the indicated antibodies.
Statistics
Statistical analyses were performed using the Statistical Program for Social Sciences (SPSS) software (Version 17.0, SPSS Inc., Chicago, IL, USA). Chi-square tests were used to assess the difference and correlation between the expression of RSK4 and the clinicopathological features of RCC patients. Both univariate and multivariate survival analyses were performed. The survival curves were constructed using a Kaplan–Meier analysis. The differences between curves were tested using the log-rank test. Cox regression analysis was used to test for the risk factors and differences in RCC-specific survival. All the experiments were done in triplicate. In cellular studies, values were expressed as mean±s.d. Comparisons between groups were assessed using the one-way ANOVA. All tests were two-sided. P-values <0.05 were considered significant.
Results
Distribution of RSK4 protein in TMA of human normal and tumour tissues
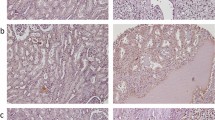
RSK4 was expressed in 11 of 30 normal human tissues. Strong positivity was seen in pancreatic ductal epithelial cells, salivary epithelial cells, sweat gland epithelial cells, and in the B lymphocytes found in the germinal centre of a tonsil (Figure 1A–D, Supplementary Table 1). The expression of RSK4 in renal tubular epithelial cells, hepatocytes, cardiomyocytes, and endometrial epithelial cells was weak (Figure 1E–H, Supplementary Table 1). RSK4 expression was seen in 14 of 50 human tumours. Clear cell RCC, uterus clear cell carcinoma, ovarian serous papillary cystadenocarcinoma, and gastric adenocarcinoma demonstrated strong positivity of RSK4 (Figure 1I–L, Supplementary Table 1), whereas some tumours manifested with weak positivity such as breast cancer and hepatocellular carcinoma.
The tissue distribution of RSK4 protein in human normal and tumour tissues. Tissue microarray is consisted of 30 human normal tissues and 54 tumour tissues, of which the expression and distribution of RSK4 are detected by immunohistochemistry with mouse anti-human monoclonal RSK4 antibody (JS-31, Santa Cruz Biotechnology). (A–D) Representative of human normal tissues with strong positivity of RSK4 protein. (A) Pancreatic ductal epithelial cells, × 400; (B) salivary epithelial cells, × 400; (C) sweat gland epithelial cells, × 400; (D) B lymphocytes in germinal centre of tonsil, × 40. (E–H) Representative of human normal tissues with weak positivity of RSK4 protein. (E) Renal tubular epithelial cells; (F) hepatocytes; (G) myocardiocytes; (H) endometrial epithelial cells, × 200; (I–L) Representative of human tumours with RSK4-positive expression. (I) Clear cell renal cell carcinoma, × 400; (J) uterus clear cell carcinoma, × 400; (K) ovarian serous papillary cystadenocarcinoma, × 200; (L) gastric adenocarcinoma, × 400.
The RSK4 protein is overexpressed in RCCs and the expression varies with different subtypes
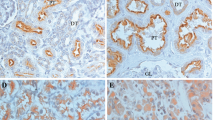
In all, 25% (5/20) normal kidney tissues expressed RSK4, whereas 61.4% (62/101) RCCs expressed RSK4. A χ2 test showed that RSK4 was overexpressed in RCCs compared with normal tissues (P=0.003). The expression of RSK4 in ccRCC, PRCC, CRCC, and other subtypes was 61.7% (29 out of 47), 47.6% (20 out of 42), 16.7% (1 out of 6), and 83.3% (5 out of 6), respectively (Figure 2, Table 1), and χ2 test showed that the difference of RSK4 expression in RCC subtypes was statistically significant (P=0.021, Table 1). Moreover, RSK4 was expressed in 14.3% (2 out of 14) of type 1 PRCC and 71.4% (20 out of 28) of type 2 PRCC (P=0.001), and the intensity of RSK4 staining was stronger in type 2 PRCC than that in type 1 PRCC (Figure 2E and F).
The expression of RSK4 protein varies in different subtypes of RCCs and is correlated with Fuhrman grade of RCC specimens. Representative of 101 cases of different RCCs subtypes are stained with RSK4 (JS-31, Santa Cruz Biotechnology) by immunohistochemistry ( × 400). (A) Fuhrman grade 1 ccRCC; (B) Fuhrman grade 2 ccRCC; (C) Fuhrman grade 3 ccRCC; (D) Fuhrman grade 4 ccRCC; (E) type 1 PRCC; (F) type 2 PRCC; (G) CRCC; (H) medullary carcinoma. Abbreviations: ccRCC=clear cell RCC; CRCC=chromophobe RCC; PRCC=papillary RCC.
The expression of RSK4 is positively correlated with high pT stage, Fuhrman grade, lymph node involvement, and distant metastasis
The expression of RSK4 increased as Fuhrman grade increased (Figure 2A–D). 28.9% (13 out of 45) of grade 1–2 RCCs and 71.4% (40 out of 56) of grade 3–4 RCCs expressed RSK4. Likewise, 21 (21 out of 61, 34.4%) stage pT1-2 tumours and 32 (32 out of 40, 80%) stage pT3-4 tumours expressed RSK4. The expression of RSK4 was significantly correlated with pT stage or Fuhrman grade (χ2 test, P<0.001, Table 1). RSK4 expression also correlated with lymph node involvement (P<0.001) and distant metastasis (P=0.039, Table 1). However, age, gender, and tumour size did not correlate with RSK4 expression (P>0.05, Table 1).
Expression of RSK4 predicts poor outcome in RCC patients
Except for some censored cases, 83 patients were followed up for survival. Among them, 38 cases were RSK4 positive and the other 45 cases were RSK4 negative in their tumours. The median survival was 97 months in patients with RSK4-negative tumours and 63 months in patients with RSK4-positive ones. Log-rank test showed that the RSK4-positive patients had a significantly worse survival than negative ones (P=0.003, Table 2). RSK4 expression was a poor prognostic indicator for patients with RCC (Figure 3A). pT stage (P<0.001), Fuhrman grade (P=0.001), lymph node involvement (P<0.001), and the presence of distant metastasis (P=0.001) were also significant prognostic indicators in a univariate analysis. Higher pT stage, higher Fuhrman grade, lymph node involvement, and distant metastasis were associated with poor survival (Table 2). Multivariate analysis revealed that RSK4 expression (HR: 2.155, P=0.042), pT stage (HR: 5.576, P<0.001), Fuhrman grade (HR: 4.118, P=0.024), lymph node involvement (HR: 7.675, P<0.001), and distant metastases (HR: 3.127, P=0.034) were independent prognostic indicators of survival (Table 2).
The positive expression of RSK4 predicts poor outcome of RCC patients. Kaplan–Meier survival curve showed the association between RSK4-positive and negative expression in 83 RCC patients (A) and therein 29 PRCC patients (B). The log-rank test demonstrated RSK4-positive expression is correlated with poor outcome (P=0.003, P=0.032).
In view of the discrepancy of RSK4 expression in different subtypes of PRCC, we further analysed the correlation of RSK4 expression and the prognosis of PRCC patients. Both uni- and multi-variate analyses showed that RSK4 expression predicts a poor outcome for PRCC patients (Figure 3B).
The expression of RSK4 in RCC fresh tissues and RCC cell lines
Real-time PCR showed that RSK4 mRNA was increased in RCCs compared with adjacent normal tissues (Figure 4A). Eight of ten RCCs had increased RSK4 protein expression in the tumour on western blot analysis (Figure 4B). RSK4 expression was higher in the RCC cell line GRC-1 and lower in ACHN (Figures 4A and B). Moreover, the two RSK4 antibodies (JS-31 and C-20, Santa Cruz Biotechnology) used for immunoblotting not only detect the wild-type RSK4 protein at 84 kDa as the dominant one, but also identified multiple bands, which were smaller such as at 48 kDa (Supplementary Figure 1).
RSK4 is overexpressed in human fresh RCC tissues and could regulate the RCC cell cycle progression via p53 pathway. (A,B) qRT-PCR (A) and western blot analysis (B) showed that the level of RSK4 mRNA and protein were higher in most RCCs compared with normal renal tissues. However, the RSK4 level varied in three RCC cell lines with higher in GRC-1 but lower in ACHN cell line. N: human normal renal tissues; T: human tumours of RCCs (RSK4 antibody for immunoblot: JS-31, Santa Cruz Biotechnology ). (C) qRT-PCR showed the RSK4 mRNA level was higher in RSK4-overexpressed ACHN cell lines and lower in RSK4-suppressed GRC-1 cell lines compared with the controls. hRSK4: ACHN cell lines transfected with human pcDNA3.1/Neo-RSK4 plasmid; vector: ACHN cell lines transfected with pcDNA3.1 vector; shRSK4: GRC-1 cell lines infected with human RSK4 shRNA viral particles by lentivirus-based transduction; scrambled: GRC-1 cell lines infected with scrambled viral particles by lentivirus-based transduction. (D) Western blot analysis confirmed the RSK4 protein level was upregulated in RSK4-overexpressed ACHN cell lines but downregulated in RSK4-suppressed GRC-1 cell lines compared with the controls (lane 1, RSK4 antibody for immunoblot: JS-31, Santa Cruz Biotechnology). RSK4-overexpressed ACHN cells showed decreased level of p53 (lane 2) but increased level of pRb (lane 4), whereas it is on the contrary in the RSK4-suppressed GRC-1 cells. However, no obvious change of the p21 (lane 3) level was observed. β-Actin was showed as loading control.
Generation of RSK4-overexpressing and RSK4-suppresed stable clones
We generated RSK4-overexpressing cell line by transfection with a pcDNA3.1/neo-RSK4 plasmid to ACHN cell line. Meanwhile, GRC-1 cells were transfected with shRSK4 lentivirus particles to decrease the expression of RSK4. The empty vector pcDNA3.1/neo and scrambled shRNA were also transfected to establish control clones. qRT-PCR and western blot analysis verified the alteration of RSK4 expression in target cells. Compared with normal and control cells, RSK4 clones showed higher RSK4 mRNA and protein expression in overexpressing RSK4 ACHN cells, and lower RSK4 expression in downregulated shRSK4 clones of GRC-1 cells (Figure 4C and D).
RSK4 could promote cell cycle progression
The cell cycle distribution in RSK4-overexpressed, RSK4-suppressed, and vector control cells was determined using flow cytometry. Compared with vector control clones, RSK4-overexpressing clones showed a significantly but slightly lower percentage of cells accumulated in the G0-G1 phase (71.08±0.51 vs 73.46±0.24, P<0.05, Supplementary Figure 2) but higher percentage of cells in the S phase (17.95±0.18 vs 15.69±0.36, P<0.05, Supplementary Figure 2). This suggested that RSK4 overexpression could promote cell cycle progression. However, there was an increase in the percentage of G0-G1 cells in the shRSK4-GRC-1 cell line (75.39±0.25 vs 59.68±0.21, P<0.05, Supplementary Figure 2) and a decrease of cells in the S and G2-M phase (19.85±0.87 vs 27.92±0.87; 4.76±0.14 vs 12.40±0.27, P<0.05, Supplementary Figure 2), indicating that inhibition of RSK4 could cause cell cycle arrest. Molecular targets involved in this effect were evaluated. RSK4-overexpressing ACHN cells demonstrated decreased p53 but increased pRb expression. No difference in p21 expression was observed, compared with control vector-transfected cells (Figure 4D). shRNA-mediated depletion of RSK4 was associated with increased expression of p53 and decreased pRb, but no marked change in p21 expression, compared with scrambled-infected cells (Figure 4D).
RSK4 enhances the invasive and metastatic ability of RCC cells by regulating the expression of MMP-9 and CD44
The ability of RSK4 to affect the invasion and migration of RCC cells in a Matrigel invasion assay was evaluated. RSK4-overexpressing ACHN cells demonstrated significantly increased invasion and migration compared with vector control cells (P<0.05), but the GRC-1 cells infected with shRSK4 manifested a significant reduction in cell invasion and migration (P<0.05, Figure 5A).
RSK4 could promote the invasive and metastatic activities of RCC cells. (A) RSK4-overexpressing ACHN cells showed elevated numbers of invasive and metastatic cells compared with normal and vector control cells (above, *P<0.05 vs normal and vector). When RSK4 was downregulated by shRNA in GRC-1 cells, the number of invasive and metastatic cells were reduced (below, *P<0.05 vs normal and scrambled). (B) RSK4-overexpressed ACHN cells showed increased level of CD44 and MMP-9; however, RSK4-suppressed GRC-1 cells manifested with decreased level of CD44 and MMP-9. β-Actin was showed as loading control (RSK4 antibody for immunoblot: JS-31, Santa Cruz Biotechnology).
To further explore the mechanisms promoting invasion and migration, the expression of CD44 and MMP-9 were evaluated. Both these factors have been reported to participate in the invasion and migration of RCCs (Cho et al, 2003; Lucin et al, 2004; Yildiz et al, 2004; Kawata et al, 2007; Lim et al, 2008). RSK4-overexpressing ACHN cells demonstrated increased expression of CD44 and MMP-9 at the protein level. Silencing of RSK4 expression in GRC-1 cells decreased expression of CD44 and MMP-9 proteins (Figure 5B). These results suggest that promotion of invasion and migration of RCC cells by RSK4 could be mediated through the regulation of CD44 and MMP-9 expression.
Specific blockade of the ERK pathway inhibits the invasiveness of RCC cells
We exposed GRC-1 cells to U0126 to block the ERK pathway or to BI-D1870 to inhibit RSK activity directly. Both inhibitors efficiently suppressed cell motility in a Transwell migration assay (P<0.05, Figure 6A). Prior treatment of cells with U0126 abolished the activation of ERK1/2 and RSK4. Treatment with the RSK inhibitor BI-D1870 (Sapkota et al, 2007) almost completely suppressed RSK4 expression without affecting the activation of ERK1/2. Simultaneously, the expression of CD44 and MMP-9 were markedly reduced in these cell lines (Figure 6B).
Specific blockade of the ERK pathway inhibits the invasiveness and metastasis of RCC cells. (A) The GRC-1 cells were pre-treated with U0126 and BI-D1870 (10 mM) for 1 h, and subsequently stimulated with fetal bovine serum (FBS) for 15 min. The blockade of RSK4 and ERK both lead to a decreased number of invasive and metastatic cells (*P<0.05 vs control). (B) Prior treatment of GRC-1 cells with U0126 abolished the activation of ERK1/2 and RSK4, whereas such treatment with the RSK inhibitor BI-D1870 almost completely suppressed RSK4 expression without affecting the activation of ERK1/2. Simutaneously, the expression of CD44 and MMP-9 were markedly reduced. β-Actin was showed as loading control (RSK4 antibody for immunoblot: JS-31, Santa Cruz Biotechnology).
Discussion
As reviewed earlier, the current knowledge on the expression and functions of RSK4 is quite limited and inconsistent. We screened the distribution of RSK4 protein in human normal and tumour tissues by immunohistochemistry. The results showed that RSK4 protein was expressed strongly in pancreas but weakly in brain, kidney, liver, and heart, which was not exactly the same as its mRNA expression reported by others (Yntema et al, 1999; Sun et al, 2013). However, transcriptional and post-transcriptional regulation of mRNA may explain this different expression between mRNA and protein level of a gene (Kozak, 2007). Moreover, RSK4 strongly positive tissues, such as salivary epithelial cells, demonstrated high proliferation, indicating that RSK4 might be a pro-proliferative factor.
RSK4 may be a tumour-suppressor gene and it has been reported to be downregulated in some tumours (ME et al, 2006; Lopez-Vicente et al, 2009; Dewdney et al, 2011). Several potential mechanisms of this downregulation exist. First, RSK4 is a component of the p53 pathway, and participates in p53-related growth arrest (Berns et al, 2004; ME et al, 2006). Second, RSK4 is thought to induce senescence via p21, contributing to cellular transformation (Lopez-Vicente et al, 2011). Third, the RSK4 promoter is hypermethylated in some cancers, leading to the absence or reduction of its expression (Dewdney et al, 2011). However, in our study, TMA screen showed RSK4 was overexpressed in RCCs and gastric carcinomas, compared with normal tissues. We subsequently detected the expression of RSK4 by immunohistochemistry in enlarged samples of normal renal tissues and RCCs, and indeed found that RSK4 was overexpressed in RCCs (P=0.003). Similar findings were observed using qRT-PCR and western blot analysis of 10 fresh RCC tumours. These findings suggest that RSK4 might be oncogenic in RCCs. Bender and Ullrich (2012) reported that overexpression of RSK4 could cause sunitinib resistance in RCC cell lines. This gives further support to a role of RSK4 in oncogenesis. Besides, RSK4 was overexpressed in breast cancer, compared with normal mammary tissues (Thakur et al, 2005, 2007). Thus, whether RSK4 is tumour promotive or suppressive is still uncertain. Recently, Sun et al (2013) reported that RSK4 protein might have several isoforms besides the known 84 kDa form. In same manner, we detected multiple forms other than the wild-type 84 kDa form, such as 48 kDa in size (Supplementary Figure 1). It is reported that there are three RSK4 mRNA transcripts at approximately 5, 6.5 and 9 kb, of which the 6.5 kb band seems to be the most prominent (Yntema et al, 1999). However, there is only one human RSK4 mRNA documented in NCBI database. Whether these protein isoforms are translated from one mRNA or other mRNA transcripts is not sure. And whether these bands are either RSK4 isoforms or RSK4 antibody reactive non-RSK4 proteins need further investigation. If RSK4 protein is proved to have other isoforms, it could explain to some extent why RSK4 acts so differently in tumour progression, as different isoforms may have different or even opposite functions, such as the roles of oestrogen receptor alpha and beta in ovarian cancer (Lazennec, 2006).
Flow cytometric analysis demonstrated that overexpression of RSK4 in RCC cells could promote cell cycle progression. To explain this result, we evaluated the expression of the cell-cycle-related proteins p53, p21, and pRb. Our results showed that overexpression of RSK4 in RCC is associated with a decrease of p53 expression and an increase of pRb, the latter of which could allow the cells through the cell cycle, leading to more cells in S and G2/M phase. In contrast, when RSK4 is inhibited by shRSK4, the increased p53 and decreased pRb caused more cells accumulated in the G0-G1 phase (Supplementary Figure 2). But surprisingly, there was no remarkable changes observed in p21 in our study. And this may be related with the sublocalisation of p21. Cmielova and Rezacova (2011) found that when in the nucleus, p21 could cause cell growth arrest, whereas in the cytoplasm, it had an anti-apoptotic effect. In RCCs, p21 could be expresssed both in the nucleus and in the cytoplasm (Weiss et al, 2007). And it is thought that overexpression of RSK4 could promote the p21 mRNA stability in the cytoplasm (Dummler et al, 2005). Thus, we consider that when RSK4 is overexpressed in our study, the nuclear p21 might be downregulated with the decrease of p53, but the cytoplasmic p21 could remain stable or even increase. And the p21 we detected by immunoblot analysis was the total proteins, irrespective of subcellular localisation, and this may explain why there was no obvious changes of p21 protein.
Approximately one-third of RCC patients present with metastasis (Lam et al, 2005). The cellular adhesion molecule CD44 and MMP-9 have important roles in this spread (Cho et al, 2003; Lucin et al, 2004; Lim et al, 2008). We found that overexpressed RSK4 was related to elevated expression of CD44 and MMP-9. When RSK4 was inhibited by shRNA, the expression of CD44 and MMP-9 decreased. Tanimura et al (2003) reported that specific blockade of the ERK pathway could inhibit the invasiveness of tumour cells by downregulating MMP-9 and CD44. As RSK4 is a downstream component of the ERK pathway, we speculate that RSK4 may act as the mediator in regulating this pro-metastatic effect in RCCs. Thus, we used the specific inhibitors U0126 and BI-D1870 to block ERK and RSK4 activity, respectively. When both or only RSK4 expression was inhibited, CD44 and MMP-9 expression and number of migratory cells decreased. These findings supported our hypothesis that RSK4 could promote RCC invasion and metastasis, which also provide a potential anti-invasive and anti-metastatic way for RCCs by specifically inhibiting RSK4.
TNM staging, Fuhrman grading, and histological subtypes are prognostic indicators for RCC patients (Bretheau et al, 1995; Moch et al, 2000; Lam et al, 2008; Lughezzani et al, 2009). However, these factors are thought to have limitations. TNM staging is composed of many factors and requires detailed information about the patient (Edge and Compton, 2010). Staging information can be missing, leading to an inaccurate classification. Fuhrman grading is more likely to be utilised for ccRCC and PRCC than for other subtypes (Fuhrman et al, 1982). It has been reported that patients with ccRCC have a worse survival than those with PRCC and CRCC, but better than those with CDC and medullary renal carcinoma (Amin et al, 2002; Cheville et al, 2003). However, Patard et al (2005) suggested that the subtype of RCC should not be considered a major prognostic variable, comparable to TNM staging and Fuhrman grading. We found that RSK4 expression in ccRCC was higher (61.7%) than that in PRCC (47.6%) and CRCC (16.7%), but lower than that in CDC and medullary renal carcinoma (83.3%; Table 1). The more aggressive the histological subtype was, the higher the positivity of RSK4 expression was. Survival analysis showed that the expression of RSK4 predicted poor prognosis in RCC patients (P=0.003, Table 2, Figure 3A). In particular, RSK4 positivity was much higher (71.4%) in type 2 PRCC than that (14.3%) in type 1 PRCC (P=0.001, Figure 3B). Its expression was related to a poor prognosis in PRCC patients, providing a new potential factor for the differential diagnosis of these two PRCC subtypes and acting as a prognostic indicator. We believe that RSK4 could be a convenient, useful, new marker for the diagnosis and prognosis of RCC patients, especially for PRCC patients.
In conclusion, the diagnosis and management of RCC is challenging. Molecular mechanisms underlying the development, growth, and metastasis of RCC are still unclear. And further studies of the molecular pathology of RCC are needed. We, for the first time, systematically evaluate the expression of RSK4 protein in different kinds of human normal and tumour tissues. The expression of RSK4 varies in different pathologic subtypes of RCC, especially in type 1 and type 2 papillary RCC. This difference could be helpful in the differential diagnosis of PRCC subtypes. Univariate and multivariate survival analysis show that RSK4 could be an independent prognostic predictor in RCC patients. Further molecular studies suggest that RSK4 could promote cell cycle progression, tumour invasion, and tumour migration in RCC cell lines, indicating that RSK4 may act as a promoter in the progression of RCC and could serve as a new potential therapeutic target for RCC patients.
Change history
03 September 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Amin MB, Tamboli P, Javidan J, Stricker H, de-Peralta Venturina M, Deshpande A, Menon M (2002) Prognostic impact of histologic subtyping of adult renal epithelial neoplasms: an experience of 405 cases. Am J Surg Pathol 26 (3): 281–291.
Athar U, Gentile TC (2008) Treatment options for metastatic renal cell carcinoma: a review. Can J Urol 15 (2): 3954–3966.
Bender C, Ullrich A (2012) PRKX, TTBK2 and RSK4 expression causes Sunitinib resistance in kidney carcinoma- and melanoma-cell lines. Int J Cancer 131 (2): E45–E55.
Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428 (6981): 431–437.
Bretheau D, Lechevallier E, de Fromont M, Sault MC, Rampal M, Coulange C (1995) Prognostic value of nuclear grade of renal cell carcinoma. Cancer 76 (12): 2543–2549.
Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML (2003) Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J Surg Pathol 27 (5): 612–624.
Cho NH, Shim HS, Rha SY, Kang SH, Hong SH, Choi YD, Hong SJ, Cho SH (2003) Increased expression of matrix metalloproteinase 9 correlates with poor prognostic variables in renal cell carcinoma. Eur Urol 44 (5): 560–566.
Cmielova J, Rezacova M (2011) p21Cip1/Waf1 protein and its function based on a subcellular localization [corrected]. J Cell Biochem 112 (12): 3502–3506.
Dewdney SB, Rimel B, Thaker PH, Thompson DM Jr., Schmidt A, Huettner P, Mutch DG, Gao F, Goodfellow PJ (2011) Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6 (RSK4) in endometrial cancers. Clin Cancer Res 17 (8): 2120–2129.
Dummler BA, Hauge C, Silber J, Yntema HG, Kruse LS, Kofoed B, Hemmings BA, Alessi DR, Frodin M (2005) Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. J Biol Chem 280 (14): 13304–13314.
Edge SB, Compton CC (2010) The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 17 (6): 1471–1474.
Fuhrman SA, Lasky LC, Limas C (1982) Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol 6 (7): 655–663.
Kawata N, Nagane Y, Hirakata H, Ichinose T, Okada Y, Yamaguchi K, Takahashi S (2007) Significant relationship of matrix metalloproteinase 9 with nuclear grade and prognostic impact of tissue inhibitor of metalloproteinase 2 for incidental clear cell renal cell carcinoma. Urology 69 (6): 1049–1053.
Kozak M (2007) Some thoughts about translational regulation: forward and backward glances. J Cell Biochem 102 (2): 280–290.
Lam JS, Klatte T, Kim HL, Patard JJ, Breda A, Zisman A, Pantuck AJ, Figlin RA (2008) Prognostic factors and selection for clinical studies of patients with kidney cancer. Crit Rev Oncol Hematol 65 (3): 235–262.
Lam JS, Leppert JT, Figlin RA, Belldegrun AS (2005) Surveillance following radical or partial nephrectomy for renal cell carcinoma. Curr Urol Rep 6 (1): 7–18.
Lazennec G (2006) Estrogen receptor beta, a possible tumor suppressor involved in ovarian carcinogenesis. Cancer Lett 231 (2): 151–157.
Lim SD, Young AN, Paner GP, Amin MB (2008) Prognostic role of CD44 cell adhesion molecule expression in primary and metastatic renal cell carcinoma: a clinicopathologic study of 125 cases. Virchows Arch 452 (1): 49–55.
Ljungberg B, Campbell SC, Choi HY, Jacqmin D, Lee JE, Weikert S, Kiemeney LA (2011) The epidemiology of renal cell carcinoma. Eur Urol 60 (4): 615–621.
Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z (2006) 2004 WHO classification of the renal tumors of the adults. Eur Urol 49 (5): 798–805.
Lopez-Vicente L, Armengol G, Pons B, Coch L, Argelaguet E, Lleonart M, Hernandez-Losa J, de Torres I, Ramon y, Cajal S (2009) Regulation of replicative and stress-induced senescence by RSK4, which is down-regulated in human tumors. Clin Cancer Res 15 (14): 4546–4553.
Lopez-Vicente L, Pons B, Coch L, Teixido C, Hernandez-Losa J, Armengol G, Ramon YCS (2011) RSK4 inhibition results in bypass of stress-induced and oncogene-induced senescence. Carcinogenesis 32 (4): 470–476.
Lucin K, Matusan K, Dordevic G, Stipic D (2004) Prognostic significance of CD44 molecule in renal cell carcinoma. Croatian Med J 45 (6): 703–708.
Lughezzani G, Capitanio U, Jeldres C, Isbarn H, Shariat SF, Arjane P, Widmer H, Perrotte P, Montorsi F, Karakiewicz PI (2009) Prognostic significance of lymph node invasion in patients with metastatic renal cell carcinoma: a population-based perspective. Cancer 115 (24): 5680–5687.
ME LL, Vidal F, Gallardo D, Diaz-Fuertes M, Rojo F, Cuatrecasas M, Lopez-Vicente L, Kondoh H, Blanco C, Carnero A, Ramon y, Cajal S (2006) New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep 16 (3): 603–608.
Moch H, Gasser T, Amin MB, Torhorst J, Sauter G, Mihatsch MJ (2000) Prognostic utility of the recently recommended histologic classification and revised TNM staging system of renal cell carcinoma: a Swiss experience with 588 tumors. Cancer 89 (3): 604–614.
Patard JJ, Leray E, Rioux-Leclercq N, Cindolo L, Ficarra V, Zisman A, De La Taille A, Tostain J, Artibani W, Abbou CC, Lobel B, Guille F, Chopin DK, Mulders PF, Wood CG, Swanson DA, Figlin RA, Belldegrun AS, Pantuck AJ (2005) Prognostic value of histologic subtypes in renal cell carcinoma: a multicenter experience. J Clin Oncol 23 (12): 2763–2771.
Sapkota GP, Cummings L, Newell FS, Armstrong C, Bain J, Frodin M, Grauert M, Hoffmann M, Schnapp G, Steegmaier M, Cohen P, Alessi DR (2007) BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J 401 (1): 29–38.
Shimizu M, Saitoh Y, Itoh H (1990) Immunohistochemical staining of Ha-ras oncogene product in normal, benign, and malignant human pancreatic tissues. Hum Pathol 21 (6): 607–612.
Sun Y, Cao S, Yang M, Wu S, Wang Z, Lin X, Song X, Liao DJ (2013) Basic anatomy and tumor biology of the RPS6KA6 gene that encodes the p90 ribosomal S6 kinase-4. Oncogene 32 (14): 1794–1810.
Tanimura S, Asato K, Fujishiro SH, Kohno M (2003) Specific blockade of the ERK pathway inhibits the invasiveness of tumor cells: down-regulation of matrix metalloproteinase-3/-9/-14 and CD44. Biochem Biophys Res Commun 304 (4): 801–806.
Thakur A, Rahman KW, Wu J, Bollig A, Biliran H, Lin X, Nassar H, Grignon DJ, Sarkar FH, Liao JD (2007) Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol Cancer Res 5 (2): 171–181.
Thakur A, Sun Y, Bollig A, Wu J, Biliran H, Banerjee S, Sarkar FH, Liao DJ (2008) Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin Cancer Res 14 (14): 4427–4436.
Thakur A, Xu H, Wang Y, Bollig A, Biliran H, Liao JD (2005) The role of X-linked genes in breast cancer. Breast Cancer Res Treat 93 (2): 135–143.
Weiss RH, Borowsky AD, Seligson D, Lin PY, Dillard-Telm L, Belldegrun AS, Figlin RA, Pantuck AD (2007) p21 is a prognostic marker for renal cell carcinoma: implications for novel therapeutic approaches. J Urol 177: q63–68, discussion 68-9.
Yildiz E, Gokce G, Kilicarslan H, Ayan S, Goze OF, Gultekin EY (2004) Prognostic value of the expression of Ki-67, CD44 and vascular endothelial growth factor, and microvessel invasion, in renal cell carcinoma. BJU Int 93 (7): 1087–1093.
Yntema HG, van den Helm B, Kissing J, van Duijnhoven G, Poppelaars F, Chelly J, Moraine C, Fryns JP, Hamel BC, Heilbronner H, Pander HJ, Brunner HG, Ropers HH, Cremers FP, van Bokhoven H (1999) A novel ribosomal S6-kinase (RSK4; RPS6KA6) is commonly deleted in patients with complex X-linked mental retardation. Genomics 62 (3): 332–343.
Acknowledgements
We thank Professor Cajal (Department of Pathology, Vall d’Hebron University Hospital, Barcelona, Spain) and Professor Jian Zhang (Department of Biochemistry and Molecular Biology, Fourth Military Medical University, Xi’an, China) for providing the human shRSK4 plasmids and the packaging plasmids. We are also grateful to the Department of Medical Records Xijing Hospital for excellent working conditions and for providing access to archival materials. The work is supported by the National Natural Science Foundation of China (No. 81001140 and No. 81272651).
Author contributions
LF carried out experiments and wrote the manuscript, PL carried out experiments, ZY analysed the data and generated the figures, GF collected the data, DJL searched the literature, YL, JZ, YZ, and LW carried out experiments, QY and YG interpreted the data, CS collected the data, GH and ZW designed the study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Fan, L., Li, P., Yin, Z. et al. Ribosomal s6 protein kinase 4: a prognostic factor for renal cell carcinoma. Br J Cancer 109, 1137–1146 (2013). https://doi.org/10.1038/bjc.2013.463
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.463
Keywords
This article is cited by
-
Integrated analysis of methylation-driven genes and pretreatment prognostic factors in patients with hepatocellular carcinoma
BMC Cancer (2021)
-
Expression of RSK4, CD44 and MMP-9 is upregulated and positively correlated in metastatic ccRCC
Diagnostic Pathology (2020)
-
Effect of RSK4 on biological characteristics of colorectal cancer
World Journal of Surgical Oncology (2018)