Abstract
Acute myocardial infarction (AMI) is a main threat to human lives worldwide. Early and accurate diagnoses warrant immediate medical care, which would reduce mortality and improve prognoses. Circulating non-coding RNAs have been demonstrated to serve as competent biomarkers for various diseases. Following the identification of cardiac-specific microRNA miR-208a in circulation, more non-coding RNAs (miR-1, miR-499 and miR-133) have been identified as biomarkers not only for the diagnosis of AMI but also for prognosis post infarction. Here, we summarized recent findings on non-coding RNAs as biomarkers for early diagnosis of ST-segment elevation myocardial infarction and for disease monitoring of myocardial infarction. In addition, the prognostic potential of non-coding RNAs in patients treated with percutaneous coronary intervention was also described. We also include studies based on biobanks, and build a miRNA release spectrum after AMI, which provides quantitative and time-lapse monitoring of AMI progress. With this spectrum, we are able to customize personal medical care, which prevents further damage. By constructing a network of circulating non-coding RNAs with high specificity and sensitivity, detailed diagnostic information was provided for personalized medicine. Unveiling the roles and kinetics of circulating non-coding RNAs may lead to a revolution in clinical diagnosis.
Similar content being viewed by others
Introduction
Over 790 000 people are diagnosed with acute myocardial infarction (AMI, often referred to as heart attack) annually in America alone1. Diagnosis as early and accurate as possible warrants immediate medical care, which would reduce mortality and improve the prognosis of AMI2,3. AMI is classified into ST-segment elevation myocardial infarction (STEMI) and non-STEMI (NSTEMI)4. For STEMI, reperfusion therapy should be administered as quickly as possible to reduce infarct size and mortality5. For NSTEMI and unstable angina (UA), another non-ST-segment elevation acute coronary syndrome, detailed revascularization strategies were recommended based on the clinical features of individual patients3. Thus, STEMI patients require early diagnosis, whereas NSTEMI and UA patients require detailed clinical features. Hence, it is critical to build a nexus of molecular biomarkers for physicians to organize an effective, personalized therapeutic schedule.
Biomarker tests combined with electrocardiographic (ECG) analysis is the main tactic for AMI diagnosis. Although ECG provide strong evidence for ischemia or infarction, ECG alone is incapable of diagnosing NSTEMI, which comprises 60%–75% of all myocardial infarctions6. In addition, both STEMI and NSTEMI demand biomarkers for final diagnosis according to European and American guidelines4,6. Biomarkers for AMI should first be quantitatively altered in AMI and thus be used to predict and monitor the pathogenic processes of AMI7,8. Meanwhile, a good biomarker should also be stable and easily accessible9. The currently preferred diagnostic biomarkers for AMI are cardiac troponin I and T (cTnI and cTnT)10. Circulating cTnI and cTnT have been the 'gold standard' of AMI diagnosis for over 20 years since they can be released from necrotic cardiomyocytes within 2–4 h post AMI10,11. Circulating cTnI and cTnT reach peak levels at 24-48 h post AMI and last for over a week12. High-sensitive cardiac troponin I and T were recently developed to improve the sensitivity and accuracy of AMI diagnosis13,14. However, false positive results with elevated cTn occurs in patients with heart failure, chronic kidney diseases and sepsis, especially in elderly patients15,16,17,18. In addition, since cTn remains in the circulation for over 7 d12, small repeat infarctions post major infarction are unlikely detectable. Thus, it is critical to identify sensitive biomarkers for extremely early diagnosis of STEMI as well as specific biomarkers that monitor the entire pathogenic processes of AMI.
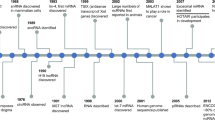
Starting with the exploration of microRNAs, numerous efforts have been made to excavate the treasures underlying non-coding RNAs (ncRNAs), which consist of over ninety percent of the human genome. MicroRNAs (miRNAs), small interference RNAs (siRNAs), long non-coding RNAs (lncRNAs) and recently identified circular RNAs (circRNAs) have all been shown to have regulatory functions or diagnostic potential in cardiovascular diseases19. MicroRNAs are endogenous short non-coding RNAs with a length of approximately 22 nt expressed in almost every cell20. Mature miRNAs bind to RNA binding proteins (RBPs) and regulate mRNA stability via recognition sites, such as AU-rich elements, in the untranslated regions of mRNA21,22,23. In 2008, scientists in the UK24, USA25 and China26 independently found that circulating microRNAs are sensitive biomarkers for cancer and other kinds of disease. Pioneering works have demonstrated that microRNAs possess huge potential as biomarkers for AMI27,28. Recently, other classes of non-coding RNAs, such as lncRNAs and circRNAs, have also been suggested as biomarkers of AMI. Despite accumulating reports on non-coding RNAs as biomarkers of AMI, we summarize recent findings and discuss future applications of non-coding RNAs as biomarkers of acute myocardial infarction with special attention to the diagnostic value and prognostic potential of these non-coding RNAs.
Non-coding RNAs as diagnostic biomarkers for AMI
MicroRNAs
The first group of miRNAs identified as biomarkers for AMI patients were miR-208a, miR-499, miR-13327 and miR-127,28. In the plasma of STEMI patients, miR-208a, miR-1, miR-499 and miR-133 were significantly increased. Among these miRNAs, miR-208a was expressed specifically in cardiomyocytes and more importantly, miR-208a showed 90.9% sensitivity and 100% specificity for AMI diagnosis, representing a more advantageous biomarker than even cTnI27. As we mentioned earlier, it is critical to identify new biomarkers for extremely early STEMI diagnosis to remedy the delayed plasma peak of cTn. Importantly, both rat models and mice models were utilized to screen for extremely early diagnostic miRNAs27,29. In rat AMI models, miR-208a increased by more than 50 fold within 1 h post coronary artery occlusion surgery and reached a peak level at 3 h after surgery, displaying kinetics that peaked just before a significant elevation in cTn27. In addition, miR-1 showed an over 200-fold increase in rat AMI models at 6 h post surgery30. A most recent study that outlines the release kinetics of circulating miR-208a in AMI patients further supported the suggestion that miR-208a is a promising diagnostic index for early diagnoses of AMI31. Even though the increase in miR-499 was not as dramatic as that of miR-208a, plasma miR-499 levels were significantly increased in rat AMI models 1 h post AMI27. A subsequent report further demonstrated that miR-208b and miR-499 were released into the circulation upon various types of myocardial damage, including AMI, and miR-208b showed an over 1600-fold increase in AMI patients32. In addition, a cohort with one thousand patients validated the increase in circulating miR-208b and miR-499 in patients with chest pain33.
The functions of miR-208a, miR-1, miR-499 and miR-133 have been well characterized in the heart34,35,36,37,38. miR-208a was initially identified in heart tissues and regulates the proper expression of β-MHC36,39. In addition, miR-208 regulates cardiomyocyte apoptosis via Bcl-2 and Bax signaling40,41. Most recently, miR-208a-3p was found to aggravate autophagy through the PDCD4-ATG5 pathway42. Recently, the diagnostic values of miR-208a, miR-1, miR-499 and miR-133 in the circulation have been intensively studied. In addition to mouse and rat AMI models, pig AMI models were established, and miR-208a, miR-1, miR-499-5p and miR-133a were all found to be elevated in pig plasma within 2 h post AMI43. Interestingly, in a pig AMI model and a rat AMI model, miR-1 was significantly increased in urine, and further analysis showed that miR-1 was also detectable in the urine of patients with AMI43,44. Recently, a study in a Chinese Han population with plasma samples drawn from patients less than 2 h after the onset of symptoms further suggested the potential of rapid diagnosis with these miRNAs45. All these reports validated that circulating miR-208a, miR-1, miR-499 and miR-133 are bona fide biomarkers for AMI. Moreover, researchers found that these cardiomyocyte-enriched miRNAs are capable of differentiating STEMI from NSTEMI. In a 444-patient cohort, miR-133a was found to distinguish STEMI, NSTEMI and UA, and miR-208a was able to distinguish STEMI from NSTEMI, whereas miR-499 and miR-208b were not able to distinguish between STEMI and NSTEMI46. However, in another cohort, miR-499 and miR-208b were found to be able to distinguish STEMI from NSTEMI47. In addition, miR-499-5p was specifically highly sensitive as a NSTEMI biomarker in elderly patients48. Additionally, miR-133a displayed distinguishable circulating levels in the serum samples of STEMI and NSTEMI patients, and serum miR-133a also showed diagnostic value for UA and takotsubo cardiomyopathy (TTC)49. More importantly, since neither ECG nor cTn is able to differentiate TTC from STEMI, it is exciting that both serum and plasma miR-133a were able to differentiate TTC from STEMI49,50. In addition, miR-499 was identified as a signature for perioperative myocardial infarction in patients undergoing coronary artery bypass graft surgery51,52. All these data indicated that the first group miRNAs identified as biomarkers for AMI possess huge potential in clinical applications, and precise interpretations of these results are promising for Precision Medicine (Figure 1).
Building a miRNA release spectrum after AMI. With current and future information on circulating miRNAs after the onset of AMI, we can build a miRNA release spectrum that contains detailed messages on when and how much miRNA is released into the circulation after AMI. With this spectrum, physicians can make extremely early diagnosis of STEMI or understand the status of an AMI patient that is either suffering from a STEMI or enduring a NSTEMI.
The number of novel miRNAs used as diagnostic biomarkers for AMI has increased rapidly. Although the sensitivity and accuracy of these circulating miRNAs vary, several novel circulating miRNAs with special release kinetics are promising candidates. Two circulating miRNAs, miR-30a and miR-195, with special kinetics post AMI were observed. These two miRNAs increased significantly at only 8 h post AMI and rapidly dropped back to baseline53. Circulating miR-122-5p also showed similar kinetics to miR-30a and miR-195, which peaked at 8 h post AMI54,55. However, circulating miR-122-5p did not show a quick drop at 12 h post AMI, but miR-30a and miR-195 did. Another circulating miRNA with unique kinetics post AMI was circulating miR-181a; its level in the circulation increased from 6 h post AMI and reached peak at 24 h post AMI56. These results suggested an option that various miRNAs with diverse releasing kinetics could be integrated together and work as monitors of the pathogenic processes of AMI. miR-328 was found increased by over 10 fold in the plasma of AMI patients in a small sample cohort57, and a large sample cohort (carried out later) further validated the over 10-fold increase of miR-328a in the plasma of AMI patients and proposed that miR-134 is another circulating miRNA that has potential in AMI diagnosis58. Although another report also suggested a rise in circulating miR-134, both studies showed that only an approximately 5-fold increase was observed post AMI58,59. Atherosclerosis-related miR-21-5p and miR-361-5-p in the circulation were increased in AMI patients as well as in patients with ischemic stroke60. Novel circulating miRNA were also identified to differentiate STEMI from NSTEMI. Circulating miR-1291 showed a significant increase in NSTEMI compared to STEMI, whereas circulating miR-663b was 2-fold higher in STEMI than in NSTEMI61. In addition, circulating miR-486 and miR-150 also varied from STEMI to NSTEMI62. Also, circulating miR-125b-5p and miR-30d-5p distinguished AMI from UA63. These data suggested that a combination of these circulating miRNAs could provide a biomarker signature to differentiate STEMI from NSTEMI and UA. Circulating miR-19a and miR-19b also showed modest increases in the plasma of AMI patients59,64. Interestingly, the circulating level of miR-22, a miRNA participating in cardiac hypertrophy65, is also elevated in the blood of AMI patients66.
Multiple novel circulating miRNAs that decreased in AMI patients have also been identified. Circulating let-7b and miR-126 were found to be dramatically decreased to 1/10 in the plasma of AMI patients from as early as 4 h post onset of AMI53,67. Moreover, the serum level of miR-126 was also decreased in AMI patients68. Since both let-7b and miR-126 play critical roles in heart development and heart diseases, it is of great importance to further investigate the biological significance of the decrease of these two circulating miRNAs69,70,71,72,73. Circulating miR-519e-5p and miR-99a were also decreased to approximately 50% in AMI patients60,74. In addition, plasma miR-145 was found to be reduced in both NSTEMI and STEMI patients and patients with heart failure75. With the fast development of high-throughput miRNA detection technologies, additional novel decreased circulating miRNAs were easily identified in AMI patients76,77,78. Circulating miR-320b and miR-125b were found to be decreased with microarray assay in the plasma of AMI patients, and these reductions were further validated in a cohort of 178 Chinese AMI patients77. Another microarray-based study showed that plasma miR-26a and miR-191 were decreased in AMI patients, which is in line with a previous report suggesting that serum miR-26a and serum miR-191 were decreased in AMI patients68,78. Notably, miR-155 and miR-126, two miRNAs critical for vascular biology69,70,79, were both decreased in the plasma of patients post off-pump coronary artery bypass graft, suggesting a potential monitoring function of miR-155 and miR-126 in cardiac surgery51. Taken together, these data suggested that by building up a thorough network of circulating miRNAs post AMI, it is possible to diagnose STEMI rapidly and monitor the pathogenic processes of AMI and thus to organize personalized therapeutic schedules for patients with a risk of AMI.
Long non-coding RNAs and circular RNAs
Recently, non-coding RNA other than miRNAs, such as long non-coding RNAs and circular RNAs, have drawn tremendous attention from scientists in the cardiovascular biology field80,81,82. Circulating forms of these non-coding RNA are also potential biomarkers for cardiovascular diseases19,83. Here, we summarize recent research on non-coding RNAs (other than miRNAs) as diagnostic biomarkers of AMI.
The first study on circulating lncRNAs tested the expression levels of five lncRNAs in the blood cells of 414 patients and 86 healthy controls84. Four out of the five lncRNAs, hypoxia inducible factor 1A antisense RNA 2 (aHIF), cyclin-dependent kinase inhibitor 2B antisense RNA 1 (ANRIL), potassium voltage-gated channel, KQT-like subfamily, member 1 opposite strand/antisense transcript 1 (KCNQ1OT1) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), are differentially expressed in AMI patients and more importantly, MALAT1, ANRIL, KCNQ1OT1 and MI associated transcript (MIAT) were able to distinguish STEMI from NSTEMI84. Two more lncRNAs, Zinc finger antisense 1 (ZFAS1) and Cdr1 antisense (CDR1AS), were identified with altered expression levels in the whole blood of AMI patients85. Another study showed alterations of over 5000 lncRNAs in the whole blood of Chinese Uyghur AMI patients86. In addition, lncRNA Urothelial carcinoma-associated 1 (UCA1), which is detectable in plasma, decreased in AMI patients at two hours after the onset of symptoms87. Although these studies provided excellent scientific significance, the sensitivity of these lncRNAs as biomarkers was not comparable to the miRNAs identified as diagnostic biomarkers for AMI. The expression of ZFAS1, CDRAS and MIAT were also affected in a MI mice model, and attenuation of MIAT abrogated the fibrogenesis induced by MI85,88. CircRNAs are a covalently closed circular form of non-coding RNA that presents in various organisms89,90. Due to their specific structure, circRNAs have great potential as biomarkers in disease diagnosis91. CircRNA Cdr1as was elevated in myocardial tissues from MI mice, suggesting that circRNAs may also have potential as diagnostic biomarkers for AMI92. In addition, a circRNA microarray was applied to detect altered circRNAs in MI mice, which further provided bases for the application of circRNAs as biomarkers93.
In summary, several attempts to use non-coding RNAs, especially miRNAs, as diagnostic biomarkers for AMI have become milestones in biomarker identification. Researchers and clinicians are paying more attention to uniting the protocols for sample collection and management. Thus, specific novel non-coding RNAs with specific diagnostic values can be expected in the future. Nonetheless, a handful of circulating miRNAs have already shown better qualities than traditional biomarkers, such as cTn. Further kinetic characterization of these miRNAs will provide detailed information for AMI diagnosis.
Non-coding RNAs as prognostic biomarkers post AMI
Non-coding RNAs as prognostic biomarkers for cardiac death post AMI
Since miRNAs are relatively stable due to various regulatory mechanisms94,95, researchers are not satisfied with using circulating miRNAs as only diagnostic biomarkers for AMI. Hence, ever since their identification, efforts have been put into using circulating miRNAs as prognostic biomarkers post AMI.
Among the first group of circulating miRNAs identified in AMI patients, miR-133a and miR-208b were first associated with mortality46. High levels of circulating miR-133a and miR-208b were found to be associated with all-cause mortality at 6 months in a 444-patient cohort46. The association between increased circulating miR-208b levels and mortality was subsequently validated by other AMI cohorts96,97. Moreover, high plasma levels of miR-499 were also associated with 30-d, 4-month, 1-year, 2-year and 6-year mortality96,97,98. Based on a miRNA array, increased levels of serum miR-155 and miR-380 were strongly associated with cardiac death99. More importantly, miR-192, miR-194 and miR-34 were significantly higher in the serum collected from a group of patients who developed ischemic heart failure after AMI onset100. In addition, high levels of serum miR-145 were also associated with cardiac death and heart failure in post AMI patients101. Recently, it was proposed that the ratio of serum miR-122-5p/133b measured at the time of cardiac catheterization was a strong predictor of cardiac death102. Although future investigations for the validation of these miRNAs as prognostic biomarkers post AMI are necessary, it is undeniable that aberrant levels of circulating miRNAs provided predictive information on adverse cardiovascular events post AMI, such as cardiac death and heart failure.
Non-coding RNAs as prognostic biomarkers for cardiac function post AMI
Despite improvements in reperfusion rates and secondary preventive medications post AMI, cardiac dysfunction, especially left ventricular dysfunction, developed at a relatively high rates103,104. Thus, it is of special importance to identify biomarkers that predict cardiac dysfunction in patients who suffered from AMI and subsequently provide medical suggestions for these patients. Based on the valuable information behind circulating non-coding RNAs in both diagnostic and prognostic levels for AMI patients, increasing studies have focused on the prognostic value of circulating non-coding RNAs for cardiac function post onset of AMI.
In addition to the roles in AMI diagnosis and cardiac death prediction post AMI, high serum levels of miR-133a were found to be associated with decreased myocardial salvage, larger infarcts and graver reperfusion injury in AMI patients post reperfusion105. In addition, elevated plasma levels of miR-133a were associated with coronary artery stenosis in patients with coronary heart disease106. Circulating miR-1, miR-208b and miR-499 were found to be negatively associated with left ventricular ejection fraction (LVEF) in patients treated with percutaneous coronary intervention96. In addition to miRNAs, high levels of circulating lncRNA MALAT1 also predicted poor LVEF in AMI patients four months post reperfusion84. Circulating lncRNA were also proposed to have roles in cardiac fibrosis post infarction88,107. Left ventricular (LV) remodeling is considered to be a predominate cause of heart failure in AMI patients108. By large-scale screening, several plasma miRNAs, especially decreased plasma miR-150, were found to be strongly associated with LV remodeling109. It was further characterized that a panel of 4 circulating miRNAs, miR-16/miR-27a/miR-101/miR-150, predicted LV remodeling after AMI with a net reclassification improvement of 66%110. In addition, increased circulating levels of miR-208b, miR-34a, miR-21 and miR-155 were all associated with LV remodeling after AMI111,112,113. Importantly, a circular RNA, MICRA, was identified recently as a circulating biomarker that predicts LV dysfunction, which expanded our understanding of circular RNAs114.
Taken together, circulating non-coding RNAs showed promising roles as prognostic biomarkers post AMI. Future validations of the previous findings in larger cohorts and standardizations of detection schemes could further promote the recognition and application of non-coding RNAs as biomarkers for risk stratification of AMI patients.
Non-coding RNAs as risk predictive biomarkers for AMI
Two studies with unique significance based on large biobanks identified circulating miRNAs that predict future myocardial infarction in healthy individuals115,116. Nineteen circulating miRNAs of 820 participants of the Bruneck cohort were quantified, and the association between levels of these circulating miRNAs and myocardial infarction incidence were estimated. In the end, researchers found that increased levels of circulating miR-126 and decreased levels of circulating miR-223 and miR-197 were associated with incidence of myocardial infarction in apparently health participants115. Based on the HUNT (The Nord-Trøndelag Health Study) report, a combination of 5 miRNAs, miR-106-5p/miR-424-5p/let-7g-5p/miR-144-3p/miR-660-5p, correctly predicted 77.6% incidence of myocardial infarction in apparently health participants. More importantly, by adding this miRNA combination to the Framingham Risk Score, the AUC increased significantly from 0.72 to 0.91116. These two illuminating studies indicated a potential use of miRNAs as biomarkers to predict heart attack for apparently healthy people. With the popularization of physical examinations and the development of personalized medicine, the establishment of miRNAs as biomarkers to predict the incidence of heart attacks is evolutionary meaningful.
Conclusion and perspectives
Former genomic 'garbage' has now proven itself as a powerful regulator in every biological process. The identification of each type of non-coding RNAs has led a revolution not only in our understanding of the molecular network but also in clinical applications. Multiple miRNAs have exhibited excellent properties as biomarkers for cancers, neurological diseases and cardiovascular diseases117,118,119,120,121. Cardiac-specific miR-208a is no doubt the most promising STEMI biomarker thus far. Not only because the specificity and sensitivity of miR-208a are outstanding but also because the kinetics of miR-208a release showed superiority over the 'gold standard' cTn: 1) miR-208a can be detected within 2 h, which is earlier than cTn, and allows extremely early diagnosis of AMI; and 2) miR-208a declined to baseline within 24 h, which enables the detection of minor cardiac events post major infarction. In addition, based on existing reports, we are able to build a miRNA release spectrum (Figure 1) after AMI, which provides quantitative and time-lapse monitoring of AMI progress. With this spectrum, we are able to customize personal medical care, which prevents further damage.
Despite what researchers have accomplished, validations of non-coding RNAs with dramatic changes and swift responses to the onset of AMI in larger cohorts are imperatively needed. Meanwhile, the establishment of standardized detection methods and release kinetics of novel non-coding RNAs are equally important. To finally facilitate the clinical applications of non-coding RNAs as biomarkers for AMI, future studies should focus on the following: 1) patient information, especially the detailed time of symptom onset, which helps identify extremely early biomarkers for STEMI; 2) inclusion of multiple examination time points, which enables long-term monitoring of the course of NSTEMI or UA; and 3) collaboration to increase cohort size and unify detection methods, which improves the applicability of certain biomarkers to a wider population. Meanwhile, it is of significance to further analyze the predictive value of non-coding RNAs for incident myocardial infarction of apparently healthy people.
Even though several animal models have been utilized to describe the accurate release kinetics of these non-coding RNAs that may serve as biomarkers for AMI, elaborate release kinetics that reflect the authentic release kinetics of AMI patients are still not available. A nonhuman primate AMI model may provide infusive results because 1) nonhuman primates are so close to human beings that they share highly conserved genetic regulation networks; thus, all kinds of non-coding RNA, even lncRNA, could be exploited. 2) As a big animal model, primates have enough blood for time-lapse sampling. Thus, it is of great potential to utilize a primate AMI model to further explore the applications of non-coding RNAs as biomarkers for AMI.
In summary, circulating non-coding RNAs, especially miRNAs, displayed numerous advantages as biomarkers and built a multi-level nexus that illustrates that the biological nature of AMI is a promising prospect for AMI diagnosis and prognosis.
References
Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, et al. Heart disease and stroke statistics-2017 update: a report from the american heart association. Circulation 2017; 135: e146–e603.
Shibata T, Kawakami S, Noguchi T, Tanaka T, Asaumi Y, Kanaya T, et al. Prevalence, clinical features, and prognosis of acute myocardial infarction attributable to coronary artery embolism. Circulation 2015; 132: 241–50.
Reed GW, Rossi JE, Cannon CP. Acute myocardial infarction. Lancet 2017; 389: 197–210.
Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD, et al. Third universal definition of myocardial infarction. Eur Heart J 2012; 33: 2551–67.
Kolh P, Windecker S. ESC/EACTS myocardial revascularization guidelines 2014. Eur Heart J 2014; 35: 3235–6.
Levine GN, Bates ER, Blankenship JC, Bailey SR, Bittl JA, Cercek B, et al. 2015 ACC/AHA/SCAI focused update on primary percutaneous coronary intervention for patients with ST-elevation myocardial infarction: an update of the 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention and the 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the american college of cardiology/american heart association task force on clinical practice guidelines and the society for cardiovascular angiography and interventions. Circulation 2016; 133: 1135–47.
Navickas R, Gal D, Laucevicius A, Taparauskaite A, Zdanyte M, Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res 2016; 111: 322–37.
Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation 2006; 113: 2335–62.
Danielson KM, Rubio R, Abderazzaq F, Das S, Wang YE. High throughput sequencing of extracellular RNA from human plasma. PLoS One 2017; 12: e0164644.
Morrow DA, Cannon CP, Jesse RL, Newby LK, Ravkilde J, Storrow AB, et al. National academy of clinical biochemistry laboratory medicine practice guidelines: Clinical characteristics and utilization of biochemical markers in acute coronary syndromes. Circulation 2007; 115: e356–75.
Babuin L, Jaffe AS. Troponin: the biomarker of choice for the detection of cardiac injury. CMAJ 2005; 173: 1191–202.
Jaffe AS, Babuin L, Apple FS. Biomarkers in acute cardiac disease: the present and the future. J Am Coll Cardiol 2006; 48: 1–11.
Roffi M, Patrono C, Collet JP, Mueller C, Valgimigli M, Andreotti F, et al. 2015 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: Task force for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation of the European society of cardiology (ESC). Eur Heart J 2016; 37: 267–315.
Keller T, Zeller T, Peetz D, Tzikas S, Roth A, Czyz E, et al. Sensitive troponin I assay in early diagnosis of acute myocardial infarction. N Engl J Med 2009; 361: 868–77.
Abbas NA, John RI, Webb MC, Kempson ME, Potter AN, Price CP, et al. Cardiac troponins and renal function in nondialysis patients with chronic kidney disease. Clin Chem 2005; 51: 2059–66.
Finsterer J, Stollberger C, Krugluger W. Cardiac and noncardiac, particularly neuromuscular, disease with troponin-T positivity. Neth J Med 2007; 65: 289–95.
Rosjo H, Varpula M, Hagve TA, Karlsson S, Ruokonen E, Pettila V, et al. Circulating high sensitivity troponin T in severe sepsis and septic shock: distribution, associated factors, and relation to outcome. Intensive Care Med 2011; 37: 77–85.
Eggers KM, Lind L, Ahlstrom H, Bjerner T, Ebeling Barbier C, Larsson A, et al. Prevalence and pathophysiological mechanisms of elevated cardiac troponin I levels in a population-based sample of elderly subjects. Eur Heart J 2008; 29: 2252–8.
Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J 2017. doi: 10.1093/eurheartj/ehx165.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97.
Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005; 120: 623–34.
Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007; 128: 1105–18.
Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008; 133: 128–41.
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008; 141: 672–5.
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105: 10513–8.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006.
Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 2010; 31: 659–66.
Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun 2010; 391: 73–7.
Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem 2009; 55: 1944–9.
Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010; 119: 87–95.
Bialek S, Gorko D, Zajkowska A, Koltowski L, Grabowski M, Stachurska A, et al. Release kinetics of circulating miRNA-208a in the early phase of myocardial infarction. Kardiol Pol 2015; 73: 613–9.
Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010; 3: 499–506.
Devaux Y, Mueller M, Haaf P, Goretti E, Twerenbold R, Zangrando J, et al. Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J Intern Med 2015; 277: 260–71.
Li Q, Song XW, Zou J, Wang GK, Kremneva E, Li XQ, et al. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci 2010; 123: 2444–52.
Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007; 100: 416–24.
Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009; 119: 2772–86.
Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007; 13: 613–8.
Matkovich SJ, Hu Y, Eschenbacher WH, Dorn LE. Dorn GW 2nd. Direct and indirect involvement of microRNA-499 in clinical and experimental cardiomyopathy. Circ Res 2012; 111: 521–31.
van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 2009; 17: 662–73.
Huang Y, Yang Y, He Y, Huang C, Meng X, Li J. MicroRNA-208a potentiates angiotensin II-triggered cardiac myoblasts apoptosis via inhibiting Nemo-like kinase (NLK). Curr Pharm Des 2016; 22: 4868–75.
Tony H, Meng K, Wu B, Yu A, Zeng Q, Yu K, et al. MicroRNA-208a dysregulates apoptosis genes expression and promotes cardiomyocyte apoptosis during ischemia and its silencing improves cardiac function after myocardial infarction. Mediators Inflamm 2015; 2015: 479123.
Wang L, Ye N, Lian X, Peng F, Zhang H, Gong H. MiR-208a-3p aggravates autophagy through the PDCD4-ATG5 pathway in Ang II-induced H9c2 cardiomyoblasts. Biomed Pharmacother 2017; 98: 1–8.
Gidlof O, Andersson P, van der Pals J, Gotberg M, Erlinge D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology 2011; 118: 217–26.
Cheng Y, Wang X, Yang J, Duan X, Yao Y, Shi X, et al. A translational study of urine miRNAs in acute myocardial infarction. J Mol Cell Cardiol 2012; 53: 668–76.
Liu X, Fan Z, Zhao T, Cao W, Zhang L, Li H, et al. Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: An independent study of Han population. Exp Gerontol 2015; 72: 230–8.
Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, et al. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol 2011; 51: 872–5.
Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, et al. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem 2012; 58: 559–67.
Olivieri F, Antonicelli R, Lorenzi M, D'Alessandra Y, Lazzarini R, Santini G, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol 2013; 167: 531–6.
Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet 2011; 4: 446–54.
Jaguszewski M, Osipova J, Ghadri JR, Napp LC, Widera C, Franke J, et al. A signature of circulating microRNAs differentiates takotsubo cardiomyopathy from acute myocardial infarction. Eur Heart J 2014; 35: 999–1006.
Pourrajab F, Torkian Velashani F, Khanaghaei M, Hekmatimoghaddam S, Rahaie M, Zare-Khormizi MR. Comparison of miRNA signature versus conventional biomarkers before and after off-pump coronary artery bypass graft. J Pharm Biomed Anal 2017; 134: 11–7.
Yao Y, Du J, Cao X, Wang Y, Huang Y, Hu S, et al. Plasma levels of microRNA-499 provide an early indication of perioperative myocardial infarction in coronary artery bypass graft patients. PLoS One 2014; 9: e104618.
Long G, Wang F, Duan Q, Yang S, Chen F, Gong W, et al. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One 2012; 7: e50926.
Li Z, Lu J, Luo Y, Li S, Chen M. High association between human circulating microRNA-497 and acute myocardial infarction. ScientificWorldJournal 2014; 2014: 931845.
Yao XL, Lu XL, Yan CY, Wan QL, Cheng GC, Li YM. Circulating miR-122-5p as a potential novel biomarker for diagnosis of acute myocardial infarction. Int J Clin Exp Pathol 2015; 8: 16014–9.
Zhu J, Yao K, Wang Q, Guo J, Shi H, Ma L, et al. Circulating miR-181a as a potential novel biomarker for diagnosis of acute myocardial infarction. Cell Physiol Biochem 2016; 40: 1591–602.
Wang R, Li N, Zhang Y, Ran Y, Pu J. Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern Med 2011; 50: 1789–95.
He F, Lv P, Zhao X, Wang X, Ma X, Meng W, et al. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol Cell Biochem 2014; 394: 137–44.
Wang KJ, Zhao X, Liu YZ, Zeng QT, Mao XB, Li SN, et al. Circulating miR-19b-3p, miR-134-5p and miR-186-5p are promising novel biomarkers for early diagnosis of acute myocardial infarction. Cell Physiol Biochem 2016; 38: 1015–29.
Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, et al. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS One 2014; 9: e105734.
Peng L, Chun-guang Q, Bei-fang L, Xue-zhi D, Zi-hao W, Yun-fu L, et al. Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn Pathol 2014; 9: 89.
Zhang R, Lan C, Pei H, Duan G, Huang L, Li L. Expression of circulating miR-486 and miR-150 in patients with acute myocardial infarction. BMC Cardiovasc Disord 2015; 15: 51.
Jia K, Shi P, Han X, Chen T, Tang H, Wang J. Diagnostic value of miR-30d-5p and miR-125b-5p in acute myocardial infarction. Mol Med Rep 2016; 14: 184–94.
Zhong J, He Y, Chen W, Shui X, Chen C, Lei W. Circulating microRNA-19a as a potential novel biomarker for diagnosis of acute myocardial infarction. Int J Mol Sci 2014; 15: 20355–64.
Xu XD, Song XW, Li Q, Wang GK, Jing Q, Qin YW. Attenuation of microRNA-22 derepressed PTEN to effectively protect rat cardiomyocytes from hypertrophy. J Cell Physiol 2012; 227: 1391–8.
Maciejak A, Kiliszek M, Opolski G, Segiet A, Matlak K, Dobrzycki S, et al. miR-22-5p revealed as a potential biomarker involved in the acute phase of myocardial infarction via profiling of circulating microRNAs. Mol Med Rep 2016; 14: 2867–75.
Long G, Wang F, Duan Q, Chen F, Yang S, Gong W, et al. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci 2012; 8: 811–8.
Hsu A, Chen SJ, Chang YS, Chen HC, Chu PH. Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed Res Int 2014; 2014: 418628.
Zou J, Li WQ, Li Q, Li XQ, Zhang JT, Liu GQ, et al. Two functional microRNA-126s repress a novel target gene p21-activated kinase 1 to regulate vascular integrity in zebrafish. Circ Res 2011; 108: 201–9.
Chen J, Zhu RF, Li FF, Liang YL, Wang C, Qin YW, et al. MicroRNA-126a directs lymphangiogenesis through interacting with chemokine and Flt4 signaling in zebrafish. Arterioscler Thromb Vasc Biol 2016; 36: 2381–93.
Chistiakov DA, Orekhov AN, Bobryshev YV. The role of miR-126 in embryonic angiogenesis, adult vascular homeostasis, and vascular repair and its alterations in atherosclerotic disease. J Mol Cell Cardiol 2016; 97: 47–55.
Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y, Zhou HH. Let-7 in cardiovascular diseases, heart development and cardiovascular differentiation from stem cells. Int J Mol Sci 2013; 14: 23086–102.
Cheng J, Zhang P, Jiang H. Let-7b-mediated pro-survival of transplanted mesenchymal stem cells for cardiac regeneration. Stem Cell Res Ther 2015; 6: 216.
Yang SY, Wang YQ, Gao HM, Wang B, He Q. The clinical value of circulating miR-99a in plasma of patients with acute myocardial infarction. Eur Rev Med Pharmacol Sci 2016; 20: 5193–7.
Zhang M, Cheng YJ, Sara JD, Liu LJ, Liu LP, Zhao X, et al. Circulating microRNA-145 is associated with acute myocardial infarction and heart failure. Chin Med J (Engl) 2017; 130: 51–6.
Liang J, Bai S, Su L, Li C, Wu J, Xia Z, et al. A subset of circulating microRNAs is expressed differently in patients with myocardial infarction. Mol Med Rep 2015; 12: 243–7.
Huang S, Chen M, Li L, He M, Hu D, Zhang X, et al. Circulating MicroRNAs and the occurrence of acute myocardial infarction in Chinese populations. Circ Cardiovasc Genet 2014; 7: 189–98.
Li C, Chen X, Huang J, Sun Q, Wang L. Clinical impact of circulating miR-26a, miR-191, and miR-208b in plasma of patients with acute myocardial infarction. Eur J Med Res 2015; 20: 58.
Tian FJ, An LN, Wang GK, Zhu JQ, Li Q, Zhang YY, et al. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc Res 2014; 103: 100–10.
Bar C, Chatterjee S, Thum T. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation 2016; 134: 1484–99.
Li M, Ding W, Sun T, Tariq MA, Xu T, Li P, et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J 2018; 285: 220–32.
Gomes CPC, Spencer H, Ford KL, Michel LYM, Baker AH, Emanueli C, et al. The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol Ther Nucleic Acids 2017; 8: 494–507.
Busch A, Eken SM, Maegdefessel L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann Transl Med 2016; 4: 236.
Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 2014; 115: 668–77.
Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep 2016; 6: 22384.
Zhai H, Li XM, Liu F, Chen BD, Zheng H, Wang XM, et al. Expression pattern of genome-scale long noncoding RNA following acute myocardial infarction in Chinese Uyghur patients. Oncotarget 2017; 8: 31449–64.
Yan Y, Zhang B, Liu N, Qi C, Xiao Y, Tian X, et al. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res Int 2016; 2016: 8079372.
Qu X, Du Y, Shu Y, Gao M, Sun F, Luo S, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep 2017; 7: 42657.
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73: 3852–6.
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014; 9: e90859.
Han YN, Xia SQ, Zhang YY, Zheng JH, Li W. Circular RNAs: A novel type of biomarker and genetic tools in cancer. Oncotarget 2017; 8: 64551–63.
Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 2016; 11: e0151753.
Guo Y, Luo F, Liu Q, Xu D. Regulatory non-coding RNAs in acute myocardial infarction. J Cell Mol Med 2017; 21: 1013–23.
Zhang Z, Qin YW, Brewer G, Jing Q. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip Rev RNA 2012; 3: 593–600.
Zhang Z, Zou J, Wang GK, Zhang JT, Huang S, Qin YW, et al. Uracils at nucleotide position 9-11 are required for the rapid turnover of miR-29 family. Nucleic Acids Res 2011; 39: 4387–95.
Gidlof O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, et al. Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord 2013; 13: 12.
Goretti E, Vausort M, Wagner DR, Devaux Y. Association between circulating microRNAs, cardiovascular risk factors and outcome in patients with acute myocardial infarction. Int J Cardiol 2013; 168: 4548–50.
Olivieri F, Antonicelli R, Spazzafumo L, Santini G, Rippo MR, Galeazzi R, et al. Admission levels of circulating miR-499-5p and risk of death in elderly patients after acute non-ST elevation myocardial infarction. Int J Cardiol 2014; 172: e276–8.
Matsumoto S, Sakata Y, Nakatani D, Suna S, Mizuno H, Shimizu M, et al. A subset of circulating microRNAs are predictive for cardiac death after discharge for acute myocardial infarction. Biochem Biophys Res Commun 2012; 427: 280–4.
Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 2013; 113: 322–6.
Dong YM, Liu XX, Wei GQ, Da YN, Cha L, Ma CS. Prediction of long-term outcome after acute myocardial infarction using circulating miR-145. Scand J Clin Lab Invest 2015; 75: 85–91.
Cortez-Dias N, Costa MC, Carrilho-Ferreira P, Silva D, Jorge C, Calisto C, et al. Circulating miR-122-5p/miR-133b ratio is a specific early prognostic biomarker in acute myocardial infarction. Circ J 2016; 80: 2183–91.
Leask A. Getting to the heart of the matter: new insights into cardiac fibrosis. Circ Res 2015; 116: 1269–76.
Savoye C, Equine O, Tricot O, Nugue O, Segrestin B, Sautiere K, et al. Left ventricular remodeling after anterior wall acute myocardial infarction in modern clinical practice (from the REmodelage VEntriculaire [REVE] study group). Am J Cardiol 2006; 98: 1144–9.
Eitel I, Adams V, Dieterich P, Fuernau G, de Waha S, Desch S, et al. Relation of circulating MicroRNA-133a concentrations with myocardial damage and clinical prognosis in ST-elevation myocardial infarction. Am Heart J 2012; 164: 706–14.
Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, et al. Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J Transl Med 2013; 11: 222.
Qu X, Song X, Yuan W, Shu Y, Wang Y, Zhao X, et al. Expression signature of lncRNAs and their potential roles in cardiac fibrosis of post-infarct mice. Biosci Rep 2016; 36. pii: e00337.
Gaasch WH, Zile MR. Left ventricular structural remodeling in health and disease: with special emphasis on volume, mass, and geometry. J Am Coll Cardiol 2011; 58: 1733–40.
Devaux Y, Vausort M, McCann GP, Zangrando J, Kelly D, Razvi N, et al. MicroRNA-150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ Cardiovasc Genet 2013; 6: 290–8.
Devaux Y, Vausort M, McCann GP, Kelly D, Collignon O, Ng LL, et al. A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS One 2013; 8: e70644.
Lv P, Zhou M, He J, Meng W, Ma X, Dong S, et al. Circulating miR-208b and miR-34a are associated with left ventricular remodeling after acute myocardial infarction. Int J Mol Sci 2014; 15: 5774–88.
Liu X, Dong Y, Chen S, Zhang G, Zhang M, Gong Y, et al. Circulating microRNA-146a and microRNA-21 predict left ventricular remodeling after ST-elevation myocardial infarction. Cardiology 2015; 132: 233–41.
Latet SC, Van Herck PL, Claeys MJ, Van Craenenbroeck AH, Haine SE, Vandendriessche TR, et al. Failed downregulation of circulating microRNA-155 in the early phase after ST elevation myocardial infarction is associated with adverse left ventricular remodeling. Cardiology 2017; 138: 91–6.
Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, et al. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol 2016; 68: 1247–8.
Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, et al. Prospective study on circulating microRNAs and risk of myocardial infarction. J Am Coll Cardiol 2012; 60: 290–9.
Bye A, Rosjo H, Nauman J, Silva GJ, Follestad T, Omland T, et al. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals - The HUNT study. J Mol Cell Cardiol 2016; 97: 162–8.
Pitkanen A, Loscher W, Vezzani A, Becker AJ, Simonato M, Lukasiuk K, et al. Advances in the development of biomarkers for epilepsy. Lancet Neurol 2016; 15: 843–56.
Bagnoli M, Canevari S, Califano D, Losito S, Maio MD, Raspagliesi F, et al. Development and validation of a microRNA-based signature (MiROvaR) to predict early relapse or progression of epithelial ovarian cancer: a cohort study. Lancet Oncol 2016; 17: 1137–46.
Patel M, Verma A, Aslam I, Pringle H, Singh B. Novel plasma microRNA biomarkers for the identification of colitis-associated carcinoma. Lancet 2015; 385: S78.
Viereck J, Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res 2017; 120: 381–99.
Barwari T, Joshi A, Mayr M. MicroRNAs in cardiovascular disease. J Am Coll Cardiol 2016; 68: 2577–84.
Acknowledgements
This work was supported in part by the National Key Research & Development Program of China (2017YFA0103700) and the National Natural Science Foundation of China (91339205, 31229002, 31271381, 81470407, and 81570208).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, C., Jing, Q. Non-coding RNAs as biomarkers for acute myocardial infarction. Acta Pharmacol Sin 39, 1110–1119 (2018). https://doi.org/10.1038/aps.2017.205
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2017.205
Keywords
This article is cited by
-
Light-harvesting iridium (III) complex-sensitized NiO photocathode for photoelectrochemical bioanalysis
Microchimica Acta (2024)
-
MiR-206 may regulate mitochondrial ROS contribute to the progression of Myocardial infarction via TREM1
BMC Cardiovascular Disorders (2023)
-
Potential diagnostic biomarkers: 6 cuproptosis- and ferroptosis-related genes linking immune infiltration in acute myocardial infarction
Genes & Immunity (2023)
-
Angiogenesis after ischemic stroke
Acta Pharmacologica Sinica (2023)
-
Traditional and novel diagnostic biomarkers for acute myocardial infarction
The Egyptian Journal of Internal Medicine (2022)