Abstract
Melanoma is the deadliest type of skin cancer. CD20+ melanoma stem cells (CSCs) are pivotal for metastasis and initiation of melanoma. Therefore, selective elimination of CD20+ melanoma CSCs represents an effective treatment to eradicate melanoma. Salinomycin has emerged as an effective drug toward various CSCs. Due to its poor solubility, its therapeutic efficacy against melanoma CSCs has never been evaluated. In order to target CD20+ melanoma CSCs, we designed salinomycin-loaded lipid-polymer nanoparticles with anti-CD20 aptamers (CD20-SA-NPs). Using a single-step nanoprecipitation method, salinomycin-loaded lipid-polymer nanoparticles (SA-NPs) were prepared, then CD20-SA-NPs were obtained through conjugation of thiolated anti-CD20 aptamers to SA-NPs via a maleimide-thiol reaction. CD20-SA-NPs displayed a small size of 96.3 nm, encapsulation efficiency higher than 60% and sustained drug release ability. The uptake of CD20-SA-NPs by CD20+ melanoma CSCs was significantly higher than that of SA-NPs and salinomycin, leading to greatly enhanced cytotoxic effects in vitro, thus the IC50 values of CD20-SA-NPs were reduced to 5.7 and 2.6 μg/mL in A375 CD+20 cells and WM266-4 CD+ cells, respectively. CD20-SA-NPs showed a selective cytotoxicity toward CD20+ melanoma CSCs, as evidenced by the best therapeutic efficacy in suppressing the formation of tumor spheres and the proportion of CD20+ cells in melanoma cell lines. In mice bearing melanoma xenografts, administration of CD20-SA-NPs (salinomycin 5 mg·kg−1·d−1, iv, for 60 d) showed a superior efficacy in inhibition of melanoma growth compared with SA-NPs and salinomycin. In conclusion, CD20 is a superior target for delivering drugs to melanoma CSCs. CD20-SA-NPs display effective delivery of salinomycin to CD20+ melanoma CSCs and represent a promising treatment for melanoma.
Similar content being viewed by others
Introduction
Melanoma, a type of cancer that develops from melanocytes, is an aggressive cutaneous cancer that is resistant to many chemotherapeutics1. Although melanoma is a rare form of skin cancer (it accounts for only approximately 1% of skin cancer), it is the deadliest type of skin cancer, as reflected by the fact that melanoma causes a large majority of skin cancer deaths2. In the last 30 years in the USA, the rate of melanoma incidence has been rising2. Thus, developing effective treatments to eradicate melanoma is urgent. Cancer stem cells (CSCs) are pivotal for cancer initiation and recurrence, and several studies have identified CD20+ melanoma CSCs. In melanoma tissues and cell lines, this phenotype possesses self-renewal, differentiation, and tumorigenicity capabilities3,4,5,6,7,8. Fang et al discovered that CD20+ melanoma CSCs could proliferate rapidly to form tumor spheres and could differentiate into different cell types6. Notably, elimination of CD20+ melanoma cells could lastingly eradicate melanoma, whereas elimination of other melanoma subpopulations could not7. Strikingly, the anti-CD20 antibody rituximab showed potential therapeutic value in a subset of patients with stage IV metastatic melanoma8. Therefore, the subpopulation of CD20+ melanoma CSCs is critical for the growth of melanoma, and selective elimination of this subpopulation represents an effective treatment to eradicate melanoma.
Salinomycin, an antibacterial therapeutic drug, has emerged as an effective drug toward various CSCs in a variety of cancers9. It was first discovered by Gupta et al who used a high-throughput screening approach that identified salinomycin as a promising drug against breast CSCs10. The anti-CSC mechanisms of salinomycin against CSCs include inhibition of autophagic flux and interference of the Wnt signaling11,12. To the best of our knowledge, the efficacy of salinomycin against melanoma CSCs has been never investigated. Thus, it is necessary to demonstrate whether salinomycin has therapeutic efficacy toward melanoma CSCs.
Before clinical application, the poor solubility of salinomycin needs to be conquered. Generally, salinomycin must be dissolved in ethanol before administration10. Nanoparticles represent a potent tool to improve the water solubility and biodistribution of salinomycin. Numerous researchers have developed nanoparticles to deliver salinomycin to CSCs that exhibit a superior therapeutic effect over free salinomycin in the treatment of CSCs13,14,15. In these studies, the nanoparticles are either polymer-based nanoparticles or liposomes13,14,15. Polymer-based nanoparticles are characterized by their superior stability during storage, high drug-loading capacity, and controlled drug-release ability, whereas their biocompatibility is not as high as that of liposomes16,17,18,19. In contrast, the biocompatibility of liposomes is good, whereas their clinical use is hampered by uncontrolled drug release and storage instability17. Thus, the development of lipid-polymer nanoparticles with controlled drug-release capability, high biocompatibility and a favorable pharmacokinetic profile is expected to overcome the disadvantages and combine the advantages of polymer-based nanoparticles and liposomes18,19.
Targeted nanoparticles with ligands have drawn much attention20. Among the ligands for targeted nanoparticles, antibodies are commonly used, but their application suffers greatly from their strong immunogenicity and high molecular weight20. Aptamers are single-stranded nucleic acids for selective binding to target molecules21. The merits of aptamers include low molecular weight, lack of immunogenicity and ready availability22,23. ACDA is an anti-CD20 DNA aptamer that binds to CD20 with stronger binding affinity than the Fab antibody fragment of Rituximab24. Therefore, we speculated that the ACDA could mediate specific and effective nanoparticle delivery to CD20+ melanoma CSCs.
In this study, we fabricated salinomycin-loaded lipid-polymer nanoparticles with ACDA (CD20-SA-NPs) to promote effective and specific salinomycin delivery to CD20+ melanoma CSCs. Due to the ACDA conjugation, CD20-SA-NPs were expected to be able to specifically target and eliminate CD20+ melanoma CSCs.
Materials and methods
Materials
1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-(maleimide (polyethylene glycol)-2000) (DSPE-PEG-Mal) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). Poly (D,L-lactide-co-glycolide) (PLGA, Mw 40 000–75 000, 50:50), salinomycin sodium (SA), basic fibroblast growth factor (b-FGF), epidermal growth factor (EGF), and all analytical-grade reagents were bought from Sigma-Aldrich (St Louis, MO, USA). Soybean lecithin was provided by Wako Pure Chemical Industries, Ltd (Osaka, Japan). The thiolated CD20 aptamers with the sequence 5′-SH-CTCCTCTGACTGTAACCACGCCGTATGTCCGAAATACGGAGAACAGCACTCATATGCAAGCCATACGCGGAGGTGCACGCGCATAGGTAGTCCAGAAGCC-3' were synthesized and provided by Ruibo Co, Ltd (Guangzhou, China) as described previously24. The FITC-labeled goat anti-mouse IgG, fetal bovine serum (FBS), B27 and ITS (insulin-transferrin-selenium), Roswell Park Memorial Institute (RPMI)-1640 medium, Trizol reagent, StemPro® Accutase® Cell Dissociation Reagent, and Pierce BCA Protein Assay Kit were bought from Thermo Fisher Scientific (Waltham, MA, USA). The Reverse Transcription System kit was bought from Promega (Madison, WI, USA). The Cell Counting Kit-8 kit (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan).
Melanoma cell lines
The human melanoma cell lines WM266-4 and A375 (American Type Culture Collection, ATCC, Manassas, VA, USA) were maintained in DMEM with 10% FBS, 25 mmol/L hydroxyethyl piperazine ethanesulfonic acid buffer, 100 μg/mL streptomycin, and 100 U/mL penicillin in a humidified atmosphere of 5% CO2 at 37 °C.
Analysis of CD20 expression using flow cytometry
After treatment with mouse anti-human CD20 monoclonal antibody (1 μg/mL) (30 min, 4 °C), the cells were treated with FITC-labeled goat anti-mouse IgG (30 min, 4 °C). The CD20 expression (the percentage of CD20+ cells) was measured with a Becton Dickinson FACScan flow cytometer (San Jose, CA, USA).
Isolation of CD20+ cells using magnetic cell sorting
The CD20+ cells were isolated from melanoma cells via magnetic cell sorting (Miltenyi Biotec) based on the protocol provided in the kit. Briefly, melanoma cells were treated with CD20 microbeads for 15 min at 4 °C. After being washed, the pellet was mixed with 500 μL of PBE buffer (PBS with 0.5% BSA and 5 mmol/L EDTA), and cells were separated using a magnetic column. The CD20+ cells were contained in the column flow and washed with PBE. Finally, the purity of the collected CD20+ cells was determined as described in the section Analysis of CD20 expression using flow cytometry.
Real-time polymerase chain reaction
Total RNA was extracted from the cells using a Trizol reagent-based approach. First-strand complementary DNA (cDNA) was reverse transcribed from the extracted RNA with a Reverse Transcription System kit. Real-time polymerase chain reaction (RT-PCR) was performed with a Light Cycler (Roche, Mannhein, Germany). The mRNA expression levels were normalized against β-actin and calculated and expressed as 2-ΔΔCT.
The tumor sphere assay
For the formation of tumor spheres, single-cell suspensions of CD20+ or CD20− melanoma cells were seeded (2000 cells/well) in six-well dishes with ultra-low adherent properties (Corning, MA, USA). The cells were maintained in stem cell culture medium (Dulbecco's modified Eagle's medium/Ham's F-12 medium (DMEM/F12), 20 ng/mL bFGF, 20 ng/mL EGF, 1× B27, and 1× ITS). The number of tumor spheres was determined under an inverted light microscope 7 d after seeding. Tumor spheres were enzymatically dissociated with StemPro® Accutase® Cell Dissociation Reagent for the culture of second-passage tumor spheres.
Animal studies
The mice were bought from the Shanghai Experimental Animal Center in the Shanghai Branch, Chinese Academy of Sciences (Shanghai, China). All procedures were approved by the Committee on Animals of the Second Military Medical University (Shanghai, China) and performed in accordance with the guidelines of the Committee on Animals of the Second Military Medical University (Shanghai, China).
The in vivo tumorigenicity assay
Severe combined immunodeficient (SCID) mice (∼20 g, 6 weeks) were bred under specific pathogen-free conditions. The in vivo tumorigenicity assay was performed in SCID mice as described below. Briefly, CD20+ or CD20− melanoma cells (2×105) were collected, mixed with growth-factor-reduced Matrigel (BD Matrigel™), and subcutaneously implanted into SCID mice. CD20+ and CD20− melanoma cells were inoculated on both sides of the same mice. The tumor formation of the cells was monitored for 60 d. The tumor volume=(width2×length)/2.
Fabrication of lipid-polymer nanoparticles
As described by Chan et al, the lipid-polymer nanoparticles were fabricated by nanoprecipitation18,19. First, salinomycin (3 mg) was dissolved in PLGA acetonitrile solution (1 mg/mL PLGA). DSPE-PEG-Mal (0.6 mg) and soybean lecithin (2 mg) were dissolved in 4% ethanol aqueous solution that was preheated to 65 °C. Then, the salinomycin solution was added into the lipid aqueous solution (1 mL/min). The mixture was shaken for 5 min and stirred for 6 h. With a dialysis tube (Spectra/pro 6 membrane, MWCO 1000), the mixture was dialyzed to PBS (pH 7.4) to remove unloaded drugs. To conjugate CD20 aptamers to nanoparticles, the nanoparticles (2 mL) were treated with 0.5 mL of CD20 aptamers (1 mg/mL) for 2 h under stirring. To remove free aptamers, ultrafiltration of the nanoparticles was performed with 100 000 NMWL Amicon® Ultra 15 mL centrifugal filter devices (Millipore, MA, USA). Blank nanoparticles were prepared in the same way without adding salinomycin. We prepared coumarin 6-loaded nanoparticles in the same way as above by adding coumarin 6. To reach the desired concentrations, the nanoparticles were concentrated by ultrafiltration with Amicon® Ultra 15 mL centrifugal filter devices (100 000 NMWL, Millipore, MA, USA).
The nanoparticles are defined as follows: CD20-SA-NPs (salinomycin-loaded lipid-polymer nanoparticles with CD20 aptamers), SA-NPs (salinomycin-loaded lipid-polymer nanoparticles), CD20-NPs (blank lipid-polymer nanoparticles with CD20 aptamers), C6-NPs (coumarin 6-loaded lipid-polymer nanoparticles), and CD20-C6-NPs (coumarin 6-loaded lipid-polymer nanoparticles with CD20 aptamers).
Characteristics of nanoparticles
The nanoparticle size and zeta potential were examined using a Zetasizer Nano-ZS instrument (Malvern, UK). The ultrastructure of nanoparticles was investigated with high-resolution transmission electron microscopy (TEM; JEOL JEM2100F, JEOL, Japan) after the samples were stained with 2% phosphotungstic acid (PTA).
High-performance liquid chromatography (HPLC, L-2000, Hitachi, Japan) with a reverse phase C-18 column (Diamonsil, 250 mm×4.6 mm, 5 μm) was utilized to measure the content of salinomycin in nanoparticles as described previously14. Briefly, 5 mg of lyophilized nanoparticles was dissolved in dichloromethane completely. After evaporation of the dichloromethane, 10 mL of methanol was added to form a sample for HPLC analysis. The mobile phase was deionized water/acetonitrile/tetrahydrofuran/phosphoric acid (10/85/5/0.01, v/v). The flow rate of the mobile phase was 1.5 mL/min. The effluent was detected at 210 nm, and the column temperature was 30 °C. Drug loading=salinomycin loaded in nanoparticles/total amount of nanoparticles. Drug encapsulation efficacy=salinomycin loaded in nanoparticles/salinomycin used in the preparation of nanoparticles. A coumarin 6 calibration curve was constructed to evaluate the drug loading in coumarin 6-loaded nanoparticles.
We evaluated the in vitro drug release of salinomycin as described below. PBS with or without 10% FBS was used as release media. Briefly, five milliliters of nanoparticle solution or free salinomycin dissolved in acetonitrile (equivalent to 5 mg of salinomycin) was placed in a dialysis tube (Spectra/pro 6 membrane, MWCO 1000) with the end sealed. Then, the tube was incubated in 1 L of release media (37 °C, a shaking speed of 100 revolutions per minute). One milliliter aliquots were withdrawn at different time points, and the salinomycin content in each aliquot was determined via HPLC as described above.
The in vitro nanoparticle stability was examined at 37 °C for 4 d. In brief, the nanoparticles were incubated with PBS, PBS with 10% FBS or 100% FBS at 37 °C. At different time points, an aliquot of nanoparticle solution was removed for size analysis.
Cellular uptake in vitro
Melanoma cells were seeded in 12-well plates at a density of 5×105 cells/well. Twelve hours later, the medium was replaced with fresh medium that contained coumarin 6-loaded nanoparticles or free coumarin 6 (25 ng/mL equivalent coumarin 6 concentration), and the cells were treated for another two hours at 37 °C. In the competitive assay, a high concentration of CD20 aptamers (50 mg/mL) was added 30 min before the treatments. The medium was discarded, and then, the cells were washed and analyzed using flow cytometry.
The salinomycin uptake of the nanoparticles was determined using the HPLC analysis. Melanoma cells were seeded in 12-well plates at a density of 5×105 cells/well. Twelve hours later, the medium was replaced with fresh medium containing free salinomycin or salinomycin-loaded nanoparticles (50 μg/mL equivalent salinomycin concentration), and the cells were treated for another 2 h at 37 °C. In the competitive assay, a high concentration of CD20 aptamers (50 mg/mL) was added 30 min before the treatments. After being collected, the cells were lysed with 0.5 mL of methanol and sonicated for 2 min. After the centrifugation of the sonicated cell lysate (20 000×g for 20 min), HPLC was used to analyze the salinomycin content in the cellular supernatant. The protein concentration in the cells was examined with a Pierce BCA Protein Assay Kit. The intracellular salinomycin uptake=the concentration of intracellular salinomycin/the concentration of cellular protein×100%.
CCK-8 assay
Melanoma cells were seeded in 96-well plates (1×104 cells/well) for 12 h. A series of concentrations of drugs were added to the cells, and cells were incubated for 72 h. Then, cell viability was measured using a CCK-8 assay.
Analysis of the CSC proportion in melanoma cells after treatments
The effect of drugs on the CSC proportion in melanoma cells was examined using the following methods. Melanoma cells (2×105/well) were seeded in 12-well plates overnight. Fresh medium containing salinomycin or salinomycin-loaded nanoparticles (5 μg/mL equivalent salinomycin concentration) was added, and the cells were incubated for another 48 h. The cells were then washed and incubated for 3 d. The proportion of CD20+ cells in the treated cells was analyzed using flow cytometry as described above. Alternatively, the cells were trypsinized and suspended in stem cell conditioned culture medium. The percentage of tumor sphere formation was defined as the number of formed tumor spheres (diameter ≥60 μm) on d 7 in the treatment group divided by the number of formed tumor spheres (diameter ≥60 μm) on d 7 in the untreated group (the percentage of the tumor sphere formation of the untreated group was used as a control and defined as 100%).
The therapeutic effect of drugs against melanoma in vivo
Briefly, 1×107 WM266-4 cells were inoculated subcutaneously on d 0 into BALB/c nude mice (6 weeks, ∼20 g). The tumors reached approximately 50 mm3 on d 10. From d 10, the mice were injected with salinomycin or nanoparticles (5 mg salinomycin/kg) via the tail vein, whereas free salinomycin (5 mg/kg) dissolved in ethanol was administered by intraperitoneal injection. From d 10, the administration was carried out once every two days for a total of 10 times. The formula (width2×length)/2 was adopted to measure the tumor volume. The effect of the drugs on the CSC proportion in melanoma tissues was measured on d 30. After the mice were euthanized, the excised tumors were chopped into small pieces. Collagenase I solution (1 mg/mL) was used to digest the tumors, and the cells were filtered through a 40-μm cell strainer and cultured overnight. The CSC proportion in the cells was analyzed according to the percentage of CD20+ cells and the tumor sphere formation assay described above. At the endpoint of this assay, the tumors were excised from the mice and weighed. The body weight of the mice was measured throughout the process.
Statistical analysis
The statistical package SPSS 13.0 (SPSS Inc, Chicago, IL, USA) was used to analyze the data in this study. A P value of <0.05 was considered statistically significant. A direct comparison between two groups was conducted using Student's non-paired t-test. A direct comparison between three or more groups was conducted with ANOVA tests.
Results
CD20+ melanoma cells have properties of CSCs
After magnetic cell sorting, the percentage of CD20+ cells was increased to >98% in melanoma cells, compared with 3%–4% of CD20+ cells in the original melanoma cell lines. To evaluate whether the CD20+ melanoma cells exhibited the characteristics of CSCs, analyses of stemness genes, tumor sphere formation, and in vivo tumor formation were performed (Figure 1). RT-PCR was used to measure the CSC-related genes in both CD20+ and CD20− cells (Figure 1A and 1B). The mRNA level of CD20 was increased by 11-fold in A375 CD20+ cells, compared with that in A375 CD133- cells. The expression of CD133, OCT4, NANOG, CD44 and NG2 was significantly increased in A375 CD20+ cells compared with A375 CD20− cells (P<0.05). In the case of WM266-4 cells, we obtained similar results.
CD20+ melanoma cells have the properties of melanoma CSCs. (A and B) RT-PCR analysis of the mRNA level of CSC-related genes in melanoma cells normalized to β-actin expression. The data are expressed as the mean±SD (n=3). (C and D) The number of tumor spheres generated from CD20+ or CD20− melanoma cells. The data are expressed as the mean±SD (n=6). (E and F) Tumor growth curve. CD20+ or CD20− melanoma cells (2×105) were implanted subcutaneously into SCID mice. The tumor volume=(width2×length)/2. The data are expressed as the mean±SD (n=6). The two groups of CD20+ and CD20− melanoma cells were compared using Student's non-paired t-test. *P<0.05; **P<0.01; ***P<0.001.
Sphere-forming assays have been utilized to evaluate self-renewal and differentiation of stem cells at the single-cell level in vitro25,26. CD20+ A375 cells formed more tumor spheres than CD20− A375 cells (P<0.01) (Figure 1C). In the case of WM266-4 cells, we obtained similar results (first-passage: P<0.01, second-passage: P<0.001) (Figure 1D).
Next, we examined whether CD20+ cells possess enhanced tumorigenic ability compared with CD20− cells (Figure 1E and 1F). The average tumor volume of CD20+ cells was significantly larger than that of CD20− cells after d 40 for A375 and d 20 for WM266-4 cells (P<0.05). At the endpoint, the average tumor volume of CD20+ A375 cells was ∼900 mm3, which was significantly larger than that of CD20− A375 cells (∼300 mm3, P<0.001). In the case of WM266-4 cells, we obtained similar results (Figure 1F).
Taken together, CD20+ melanoma cells showed stronger potential for self-renewal and tumorigenicity than CD20− melanoma cells, suggesting that CD20+ cells possess the characteristics of melanoma CSCs.
Characteristics of nanoparticles
As shown in Figure 2A, salinomycin-loaded lipid-polymer nanoparticles (SA-NPs) were prepared through a single-step process. Subsequently, CD20-SA-NPs were developed by conjugating thiolated CD20 aptamers to SA-NPs via a maleimide-thiol reaction. The nanoparticles characteristics are shown in Table 1. The nanoparticles were smaller than 100 nm and had a small PDI of <0.2 (suggesting a homogenous size distribution). Due to the coating of lecithin and PEG, the nanoparticles exhibited a negative zeta potential of about -20 mv. The nanoparticles had an encapsulation efficiency (EE) higher than 60%, and their drug loading was higher than 7%, suggesting that nanoprecipitation is an efficient way to load salinomycin in nanoparticles. As shown by TEM, the nanoparticles exhibited a spherical shape and a monodisperse pattern (Figure 2B). In vitro drug release was examined, and the results are shown in Figure 2C. Free salinomycin showed a rapid initial burst release (over 80% was released in 10 h). An initial burst release of ∼70% was observed in both nanoparticles in 48 h, and the cumulative drug release reached ∼80% in the following 96 h. For both nanoparticles, the drug release after 24 h was significantly higher in PBS with 10% FBS than in PBS (P<0.05). A stability assay was carried out, and the results are shown in Figure 3. Although the size of the nanoparticles was increased by approximately 15 nm at d 4, the nanoparticle size was stable throughout the 4 d, indicating that the stability of the nanoparticles was not significantly affected by the binding of plasma protein.
The development, morphology and drug release of nanoparticles. (A) Preparation of nanoparticles using the nanoprecipitation-based approach. (B) Transmission electron microscopy (TEM) image. Bars represent 100 nm and 250 nm. (C) The cumulative salinomycin release from free salinomycin or the nanoparticles in PBS or PBS with 10% FBS. The data are expressed as the mean±SD (n=3).
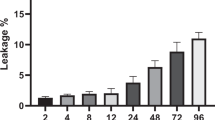
The in vitro stability assay. The data are expressed as the mean±SD (n=3).
In vitro cellular uptake
As shown in Figure 4A, C6-NPs showed a cellular uptake similar to that of coumarin-6 in A375 CD20+ cells, as reflected by their similar mean fluorescence intensity (MFI). Notably, the MFI of CD20-C6-NPs was significantly higher than that of C6-NPs (P<0.001), whereas the MFI of CD20-C6-NPs was significantly decreased after pretreatment with CD20 aptamers (P<0.01). In contrast, the MFI among coumarin-6, C6-NPs, and CD20-C6-NPs did not differ significantly in A375 CD20− cells. Similar results were obtained with WM266-4 cells. Taken together, the data suggested that CD20 aptamers could efficiently and specifically increase the uptake of CD20-C6-NPs in A375 and WM266-4 CD20+ cells but not in CD20− cells.
In vitro cellular uptake of nanoparticles in both CD20+ and CD20− melanoma cells. (A) In vitro cellular uptake of nanoparticles evaluated by flow cytometry. (B) In vitro cellular uptake of nanoparticles evaluated by HPLC. The intracellular salinomycin uptake=the concentration of intracellular salinomycin/the concentration of cellular protein×100%. In the competitive assay, a high concentration of CD20 aptamers (50 mg/mL) was added before treatments. The two results among the groups were compared using one-way ANOVA with Newman Keuls post-test. *P<0.05; **P<0.01; ***P<0.001; ns represents not significant (P>0.05). The data are expressed as the mean±SD (n=3).
Intracellular salinomycin uptake of nanoparticles was quantified using HPLC (Figure 4B). In A375 CD20+ cells, the intracellular uptake of salinomycin in CD20-SA-NPs was 7.8 μg/mg, significantly higher than in SA-NPs (2.8 μg/mg, P<0.01). Once again, the intracellular uptake of salinomycin in CD20-SA-NPs was decreased to 5.0 μg/mg after pretreatment with CD20 aptamers (P<0.05). However, the intracellular salinomycin uptake among salinomycin, SA-NPs, and CD20-SA-NPs did not differ significantly in A375 CD20− cells. Similar results were obtained with WM266-4 cells.
Taken together, CD20 aptamers significantly increased the cellular uptake of nanoparticles in CD20+ but not CD20− melanoma cells.
The cytotoxic effects of salinomycin and nanoparticles
The cytotoxic effects of salinomycin and nanoparticles were examined in CD20+ and CD20− melanoma cells (Figure 5). CD20-NPs, blank lipid-polymer nanoparticles with CD20 aptamers, did not show significant cytotoxic effects toward melanoma cells, suggesting that they have good biocompatibility. Salinomycin, SA-NPs, and CD20-SA-NPs showed a dose-dependent cytotoxicity toward melanoma cells. The IC50 values of the drugs are shown in Table 2. The IC50 of salinomycin in A375 CD20+ cells (12.9 μg/mL) was significantly lower in comparison to that in A375 CD20− cells (26.1 μg/mL) (P<0.05), suggesting that salinomycin preferably eliminates melanoma CSCs. Notably, for A375 CD20+ cells, the IC50 of CD20-SA-NPs (5.7 μg/mL) was significantly lower in comparison to that of SA-NPs (16.7 μg/mL) (P<0.01) and salinomycin (12.9 μg/mL) (P<0.01). In A375 CD20− cells, the IC50 of CD20-SA-NPs, SA-NPs, and salinomycin did not differ significantly. Further, CD20-SA-NPs showed significantly enhanced cytotoxic effects toward CD20+ cells in comparison to CD20− cells in A375 cells (5.7 μg/mL vs 30.1 μg/mL, P<0.001).
Cell proliferation after the cells were treated with the nanoparticles or salinomycin for 72 h, evaluated using a CCK-8 assay. (A) A375 CD20+; (B) A375 CD20-; (C) WM266-4 CD20+; (D) WM266-4 CD20-. The data are expressed as the mean±SD (n=3).
In the case of WM266-4 cells, we obtained similar results. The IC50 of salinomycin in WM266-4 CD20+ cells (7.5 μg/mL) was significantly lower in comparison to WM266-4 CD20− cells (18.9 μg/mL) (P<0.05). The IC50 of CD20-SA-NPs in WM266-4 CD20+ cells (2.6 μg/mL) was significantly lower in comparison to SA-NPs (11.5 μg/mL, P<0.01) and salinomycin (7.45 μg/mL, P<0.05). No significant differences in the IC50 values were found among CD20-SA-NPs, SA-NPs, and salinomycin in WM266-4 CD20− cells.
In summary, these data suggested that CD20-SA-NPs exhibited increased cytotoxic effects toward CD20+ melanoma cells compared with SA-NPs and salinomycin and enhanced cytotoxic effects toward melanoma CSCs compared with melanoma cells.
Analysis of the CSC proportion in melanoma cells after treatments
The percentage of tumor sphere formation is regarded to be associated with the percentage of CSCs13,14. Figure 6A and 6C showed that CD20-NPs did not affect the percentage of A375 tumor spheres, whereas CD20-SA-NPs decreased the number of A375 tumor spheres by 4-fold in comparison to the untreated control. Compared with salinomycin and SA-NPs, treatment with CD20-SA-NPs resulted in far fewer tumor spheres (P<0.05). In the case of WM266-4 cells, we obtained similar results (Figure 6B and 6C). CD20-SA-NPs reduced the number of WM266-4 tumor spheres by 4-fold in comparison to the untreated control and led to the formation of far fewer tumor spheres in comparison to salinomycin (P<0.05) and SA-NPs (P<0.05). Consistently, the proportion of CD20+ cells was significantly decreased by CD20-SA-NPs in A375 cells in comparison to salinomycin (P<0.05) and SA-NPs (P<0.01) (Figure 7A), and the proportion of CD20+ cells was significantly decreased by CD20-SA-NPs in WM266-4 cells in comparison to salinomycin (P<0.05) and SA-NPs (P<0.01) (Figure 7B). Thus, CD20-SA-NPs preferably target and eliminate melanoma CSCs in comparison to SA-NPs and salinomycin.
The effect of drugs on the proportion of CSCs in (A) A375 and (B) WM266-4 cells, as reflected by the percentage of tumor sphere formation. Representative images of tumor spheres formed are shown in (C) and (D). The two results among the five groups are compared by one-way ANOVA with the Newman-Keuls post-test; *P<0.05; **P<0.01; ***P<0.001; ns represents not significant (P>0.05). Data are expressed as the mean±SD (n=4).
The effect of drugs on the proportion of CSCs within the melanoma cell lines, as reflected by the proportion of CD20+ cells. The two results among the five groups were compared using one-way ANOVA with Newman-Keuls post-test; *P<0.05; **P<0.01; ***P<0.001; ns represents not significant (P>0.05). The data are expressed as the mean±SD (n=4).
The therapeutic effect of drugs against melanoma in vivo
In BALB/c nude mice bearing WM266-4 tumors, we did not observe any CD20-NP antitumor activity, indicating that blank lipid-polymer nanoparticles with CD20 aptamers could not cause tumor retardation due to lack of salinomycin (Figure 8A). Other formulations, including salinomycin, SA-NPs, and CD20-SA-NPs, showed significant therapeutic efficacy. On d 45, in comparison to other groups, the tumor volume of the CD20-SA-NP-treated group was significantly smaller (CD20-SA-NPs vs SA-NPs: P<0.01, CD20-SA-NPs vs salinomycin and CD20-NPs, and saline: P<0.001) (Figure 8A). None of the nanoparticle-treated mice showed significant changes in weight compared with the saline-treated mice, whereas the weight of the salinomycin-treated mice was lower than that of the saline-treated mice at d 28, 35, and 42 (P<0.05) (Figure 8B). The excised tumors were also weighed and compared (Figure 8C and 8D). The tumor weights in the CD20-SA-NP-treated group were significantly lower than those in other groups (CD20-SA-NPs vs SA-NPs and salinomycin: P<0.01, CD20-SA-NPs vs CD20-NPs and saline: P<0.001).
The in vivo antitumor efficacy of salinomycin and nanoparticles in a subcutaneous melanoma model. Therapy was administered once every two days (indicated by arrows) for a total of 10 times. Tumor volume=(width2×length)/2. (A) The curve of tumor growth. (B) The body weights of the mice. (C) The excised tumors. (D) The excised tumors were weighed at the experiment endpoint. On d 30, the effect of the drugs on the proportion of CSCs in WM266-4 tumors in vivo was evaluated by examining (E) the percentage of tumor sphere formation (F) and the proportion of CD20+ cells in the excised tumors. The two results among the groups were compared using one-way ANOVA with Newman-Keuls post-test. *P<0.05; **P<0.01; ***P<0.001; ns represents not significant (P>0.05). The data are expressed as the mean±SD (n=8).
The percentage of tumor sphere formation and CD20+ cells in the excised tumors were examined at the experiment endpoint (Figure 8E and 8F). Expectedly, CD20-NPs barely affected the tumor sphere formation of WM266-4 cells relative to the saline-treated control. It is noteworthy that CD20-SA-NPs significantly decreased the percentage of tumor sphere formation of WM266-4 cells compared with the SA-NPs (P<0.05) and salinomycin (P<0.05). The proportion of CD20+ melanoma cells was also evaluated in the excised tumors (Figure 8F). Compared with salinomycin and SA-NPs, CD20-SA-NPs reduced the proportion of CD20+ cells more efficiently (P<0.05), and the relative proportion of the CD20+ cells was 3-fold lower after treatment with CD20-SA-NPs compared with saline. Taken together, CD20-SA-NPs represent an effective treatment to decrease the population of melanoma CSCs in vivo.
Discussion
Melanoma is the deadliest type of skin cancer, and thus, treatment of melanoma is an urgent task for public health. Targeting melanoma CSCs represents an effective treatment to eradicate melanoma. Taking advantage of CD20 as the marker of melanoma CSCs, we utilized ACDA, an anti-CD20 aptamer, to facilitate specific and effective delivery of salinomycin-loaded nanoparticles to CD20+ melanoma CSCs. CD20-SA-NPs were demonstrated to target and eliminate CD20+ melanoma CSCs, resulting in superior therapeutic efficacy against melanoma.
Salinomycin is a promising drug against various CSCs in various types of cancer9. Not only did salinomycin show potent anti-CSC activity in several preclinical experimental studies, including in a variety of cancer cell lines and in mice bearing human cancer xenografts9, it has been tested in a few clinical pilots, which revealed that salinomycin could eliminate CSCs and induce regression of therapy-resistant cancers11. However, whether salinomycin could target melanoma CSCs has been never investigated. In this study, in two melanoma cell lines (A375 and WM266-4), the IC50 of salinomycin in CD20+ melanoma cells was significantly lower than that in CD20− melanoma cells. Moreover, salinomycin significantly decreased the formation of tumor spheres and the proportion of CD20+ cells in the two melanoma cell lines. These results suggested that salinomycin preferably eliminates melanoma CSCs. To the best of our knowledge, this is the first study reporting that salinomycin could target melanoma CSCs. Due to sustained release of salinomycin from the nanoparticles, salinomycin-loaded nanoparticles did not completely release salinomycin in the culture plates over a short period (48 h), resulting in suboptimal efficacy when tested in vitro. In contrast, when tested in vivo, salinomycin-loaded nanoparticles showed better pharmacokinetics and increased accumulation in tumors than free salinomycin, resulting in better efficacy compared with salinomycin.
CD20 is considered a superior target in the treatment of melanoma CSCs. First, only a few number of tissues (B-cells and B-cell derived malignancies) have been found to express CD20, whereas other normal tissues do not express CD2027. In contrast, other CSC markers, such as CD133, have been found to be widely expressed in normal tissues, resulting in potential damage to normal tissues if using these CSC markers as a target28. Second, since CD20 expression is restricted to B-cells (B-cells are not essential for maintenance of survival), depletion of B-cells by targeting CD20 is not supposed to induce major host toxicity29,30. Third, CD20 is a cell-surface protein that is not shed or secreted into the blood circulation, making it a stationary and accessible target. Our data showed that the superior targeting of CD20-SA-NPs to CD20+ melanoma cells is attributed to the anti-CD20 aptamers. As demonstrated by flow cytometry and HPLC assays, the uptake of CD20-SA-NPs in CD20+ melanoma cells was significantly higher compared with that of SA-NPs and salinomycin and was significantly decreased after pretreatment with anti-CD20 aptamers, indicating that anti-CD20 aptamers mediated the increased uptake of CD20-SA-NPs in CD20+ melanoma cells. Further, the IC50 of CD20-SA-NPs in CD20+ melanoma cells was significantly lower than that of SA-NPs and salinomycin, whereas the IC50 of CD20-SA-NPs in CD20− melanoma cells did not differ significantly from that of SA-NPs and salinomycin. Significantly, CD20-SA-NPs showed the best therapeutic efficacy in decreasing the formation of tumor spheres and the proportion of CD20+ cells in the melanoma cells. These data firmly demonstrated the selective toxicity of CD20-SA-NPs toward CD20+ melanoma CSCs. Further, the results showed that CD20-NPs did not affect the percentage of CD20+ cells in the melanoma cell lines and showed no antitumor activity in the in vivo antitumor assay. Thus, the CD20 aptamer alone could not eradicate melanoma CSCs.
The CD20-SA-NPs were composed of soybean lecithin, PLGA, PEGylated lipid, salinomycin, and nucleic acid aptamers. The soybean lecithin, PLGA, and PEGylated lipid are all commercially available and are US Food and Drug Administration (FDA)-approved materials. Salinomycin has already been tested in a pilot clinical trial and showed no severe acute or long-term side effects11. The application of aptamers, which are promising targeting ligands, has been rapidly increasing in the field of molecular therapy. It is notable that a vascular endothelial growth factor (VEGF) RNA aptamer, pegaptanib sodium, has been approved by the FDA, suggesting that future application of aptamers is promising31. It is well established that efficient uptake of nanoparticles in the reticuloendothelial system (RES) occurs when nanoparticles are injected into the blood17. However, our nanoparticles were PEGylated, and PEGylation significantly decreases the uptake of nanoparticles by the RES, thus reducing the potential toxicity to RES organs. Thus, CD20-SA-NPs are expected to possess superior safety in the clinic, which will surely facilitate their clinical translation.
Administration of CD20-SA-NPs in mice bearing WM266-4 melanoma xenografts showed superior efficacy in inhibition of tumor growth compared with SA-NPs and salinomycin. Notably, CD20-SA-NPs did not achieve complete elimination of melanoma. We hypothesize that the reason for this may be explained as follows. Tumors may possess distinct types of CSCs, and thus, elimination of only one CSC subpopulation may not eradicate cancer32. For melanoma, CD20, CD133, and side populations are all considered to be markers of melanoma CSCs5. Second, the conversion to CSCs by non-CSCs usually occurs due to the unstable traits of the CSC phenotype3. Thus, only killing CSCs is not sufficient to eliminate melanoma. As demonstrated by the in vitro cytotoxicity assay, our prepared CD20-SA-NPs could eliminate melanoma CSCs and non-CSCs, because the released salinomycin from the CD20-SA-NPs could effectively kill melanoma cells.
The antitumor mechanism of the CD20-SA-NPs can be elucidated as follows. After CD20-SA-NPs efficiently accumulated in melanoma tissue, they utilized the enhanced permeability and retention (EPR) effect to effectively bind and kill CD20+ melanoma cells. Although the CD20+ proportion only accounts for 3%–4% of melanoma cells, the reason why CD20-SA-NPs exhibited significantly higher efficacy than SA-NPs is that CD20-SA-NPs could eliminate the CD20+ cells, which are considered to be pivotal in the growth of cancer cells. At the same time, salinomycin gradually released from the CD20-SA-NPs effectively killed non-CSCs.
There is a limitation in this study. A comparison with CD20 antibody may better reveal the targeting efficiency of the CD20 aptamer. However, in the present study, we mainly focused on the targeting efficacy of CD20 aptamers, and the development of nanoparticles with CD20 antibody is a complex, laborious, and very costly procedure. Further, the conjugation efficiency is different between CD20 aptamers and CD20 antibody, making the comparison difficult to perform. Thus, we could not perform that experiment with CD20 antibody. However, we fully recognize the importance of the experiment with CD20 antibody and plan to perform that experiment in a future study if possible.
In conclusion, CD20 is a superior target for delivering drugs to melanoma CSCs. Thus, it is viable to use CD20 to mediate effective drug delivery to melanoma CSCs. CD20-SA-NPs realized effective salinomycin delivery to CD20+ melanoma CSCs and can potentially increase the therapeutic efficacy of salinomycin against melanoma.
Author contribution
Wu-qing WANG and Ying-ying ZHANG designed the studies; Yi-bin ZENG, Zuo-chong YU, Yan-ni HE, Tong ZHANG, Ling-bo DU, Yin-mei DONG, and Huai-wen CHEN performed the experiments; Yi-bin ZENG and Zuo-chong YU wrote the manuscript; Wu-qing WANG and Ying-ying ZHANG supervised the studies and edited the manuscript. The final manuscript has been read and approved by all authors.
References
Wong DJ, Ribas A . Targeted therapy for melanoma. Cancer Treat Res 2016; 167: 251–62.
Siegel RL, Miller KD, Jemal A . Cancer statistics. CA Cancer J Clin 2016; 66: 290–308.
Gao J, Li W, Guo Y, Feng SS . Nanomedicine strategies for sustained, controlled and targeted treatment of cancer stem cells. Nanomedicine (Lond) 2016; 11: 3261–82.
Xie F, Xu W, Yin C, Zhang G, Zhong Y, Gao J . Nanomedicine strategies for sustained, controlled, and targeted treatment of cancer stem cells of the digestive system. World J Gastrointest Oncol 2016; 8: 735–44.
Zabierowski SE, Herlyn M . Melanoma stem cells: the dark seed of melanoma. J Clin Oncol 2008; 26: 2890–4.
Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res 2005; 65: 9328–37.
Schmidt P, Kopecky C, Hombach A, Zigrino P, Mauch C, Abken H . Eradication of melanomas by targeted elimination of a minor subset of tumor cells. Proc Natl Acad Sci U S A 2011; 108: 2474–9.
Pinc A, Somasundaram R, Wagner C, Hormann M, Karanikas G, Jalili A, et al. Targeting CD20 in melanoma patients at high risk of disease recurrence. Mol Ther 2012; 20: 1056–62.
Resham K, Patel PN, Thummuri D, Guntuku L, Shah V, Bambal RB, et al. Preclinical drug metabolism and pharmacokinetics of salinomycin, a potential candidate for targeting human cancer stem cells. Chem Biol Interact 2015; 240: 146–52.
Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009; 138: 645–59.
Naujokat C, Steinhart R . Salinomycin as a drug for targeting human cancer stem cells. J Biomed Biotechnol 2012; 2012: 950658.
Yue W, Hamaï A, Tonelli G, Bauvy C, Nicolas V, Tharinger H, et al. Inhibition of the autophagic flux by salinomycin in melanoma stem-like/progenitor cells interferes with their maintenance. Autophagy 2013; 9: 714–29.
Mao X, Liu J, Gong Z, Zhang H, Liu Y, Zou H, et al. iRGD-conjugated DSPE-PEG2000 nanomicelles for targeted delivery of salinomycin for treatment of both liver cancer cells and cancer stem cells. Nanomedicine (Lond) 2015; 10: 2677–95.
Xie F, Zhang S, Liu J, Yang K, Zhang H, Liu Y, et al. Codelivery of salinomycin and chloroquine by liposomes enables synergistic antitumor activity in vitro. Nanomedicine (Lond) 2016; 11: 1831–46.
Wang M, Xie F, Wen X, Chen H, Zhang H, Liu J, et al. Therapeutic PEG-ceramide nanomicelles synergize with salinomycin to target both liver cancer cells and cancer stem cells. Nanomedicine (Lond) 2017; 12: 1025–42.
Liu Y, Pan J, Feng SS . Nanoparticles of lipid monolayer shell and biodegradable polymer core for controlled release of paclitaxel: effects of surfactants on particles size, characteristics and in vitro performance. Int J Pharm 2010; 395: 243–50.
Gao J, Chen H, Song H, Su X, Niu F, Li W, et al. Antibody-targeted immunoliposomes for cancer treatment. Mini Rev Med Chem 2013; 13: 2026–35.
Gao J, Xia Y, Chen H, Yu Y, Song J, Li W, et al. Lipid-polymer nanoparticles conjugated anti-EGFR antibody for targeted drug delivery to hepatocellular carcinoma. Nanomedicine (Lond) 2014; 9: 279–93.
Chan JM, Zhang L, Yuet KP, Liao G, Rhee JW, Langer R, et al. PLGA-lecithin-PEG core-shell nanoparticles for controlled drug delivery. Biomaterials 2009; 30: 1627–34.
Gao J, Feng SS, Guo Y . Antibody engineering promotes nanomedicine for cancer treatment. Nanomedicine (Lond) 2010; 5: 1141–5.
Nutiu R, Li Y . Structure-switching signaling aptamers. J Am Chem Soc 2003; 125: 4771–8.
Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R . Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res 2004; 64: 7668–72.
Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A 2006; 103: 6315–20.
Wu C, Wan W, Zhu J, Jin H, Zhao TJ, Li HF . Induction of potent apoptosis by an anti-CD20 aptamer via the crosslink of membrane CD20 on non-Hodgkin's lymphoma cells. Rsc Advances 2017; 7: 5158–66.
Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A 2009; 106: 16281–6.
Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008; 15: 504–14.
Maloney DG . Anti-CD20 antibody therapy for B-cell lymphomas. N Engl J Med 2012; 366: 2008–16.
Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB . Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov 2009; 8: 806–23.
McLaughlin P, Grillo-López AJ, Link BK, Levy R, Czuczman MS, Williams ME, et al. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: half of patients respond to a four-dose treatment program. J Clin Oncol 1998; 16: 2825–33.
Coiffier B, Haioun C, Ketterer N, Engert A, Tilly H, Ma D, et al. Rituximab (anti-CD20 monoclonal antibody) for the treatment of patients with relapsing or refractory aggressive lymphoma: a multicenter phase II study. Blood 1998; 92: 1927–32.
Ng EW, Shima DT, Calias P . Cunningham ET Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 2006; 5: 123–32.
Visvader JE, Lindeman GJ . Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10: 717–28.
Acknowledgements
This work was supported by 2017 Shanghai Minhang District Central Hospital Project (2017MHJC01), Fudan University Affiliated Jinshan Hospital Excellent Youth Talent Training Program (2015-2), Shanghai Jinshan District Health Family Planning System Excellent Youth Talent Training Program (JSYQ201605), and the National Natural Science Foundation of China (81302363, 81771964, and 81602358). We thank Sunlipo Biotech Research Center for Nanomedicine (Shanghai, China) for providing technical support in the preparation of nanoparticles.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zeng, Yb., Yu, Zc., He, Yn. et al. Salinomycin-loaded lipid-polymer nanoparticles with anti-CD20 aptamers selectively suppress human CD20+ melanoma stem cells. Acta Pharmacol Sin 39, 261–274 (2018). https://doi.org/10.1038/aps.2017.166
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2017.166