Abstract
Aim:
Nanobody is an antibody fragment consisting of a single monomeric variable antibody domain, which can be used for a variety of biotechnological and therapeutic purposes. The aim of this work was to isolate and characterize a human signal domain antibody against VEGFR-2 domain3 (VEGFR D3) from a phage display library.
Methods:
To produce antigen-specific recombinant nanobodies with high affinity to VEGFR2 D3, a liquid phase panning strategy was used for all rounds of panning. For nanobody expression and purification, four VEGFR2 D3-blocking clones were subcloned into a pETduet-biotin-MBP expression vector. The recombinant proteins carried an MBP tag to facilitate purification by affinity chromatography. Recombinant NTV(1–4) was obtained after an additional gel filtration chromatography step. The interactions between VEGFR2 D3 and NTV(1–4) were assessed with luminescence-based AlphaScreen assay and SPR assay. Anti-angiogenesis effects were examined in human umbilical vein endothelial cells (HUVECs).
Results:
In the AlphaScreen assay, NTV1 (100 and 200 nmol/L) elicited the highest binding signal with VEGFR2 D3; NTV2 showed moderate interactions with VEGFR2 D3; NTV3 and NTV4 exhibited little or no interaction with VEGFR2 D3. In the SPR assay, NTV1 displayed a high affinity for VEGFR2 D3 with an equilibrium dissociation constant (KD) of 49±1.8 nmol/L. NTV1 (1–1000 nmol/L) dose-dependently inhibited the proliferation of HUVECs and the endothelial tube formation by the HUVECs.
Conclusion:
The nanobody NTV1 is a potential therapeutic candidate for blocking VEGFR2. This study provides a novel and promising strategy for development of VEGFR2-targeted nanobody-based cancer therapeutics.
Similar content being viewed by others
Introduction
Angiogenesis is the process through which new blood vessels form from pre-existing vessels. This tightly regulated process plays a critical role in a variety of physiological events, including wound healing and granulation tissue formation1. However, pathological angiogenesis contributes to multiple diseases, including cancer, psoriasis, age-related macular degeneration, arthritis and blindness1,2,3,4,5. Vascular endothelial growth factors (VEGFs) are crucial regulators of vascular development during angiogenesis. In mammals, five VEGF ligands have been identified to date. These ligands bind in an overlapping pattern to three receptor tyrosine kinases, known as VEGF receptor-1, -2 and -3 (VEGFR1–3), as well as to co-receptors, such as heparan sulfate proteoglycans and neuropilins3. Of these, both VEGFR-1 and VEGFR-2 are expressed predominantly on tumor endothelial cells, indicating that VEGF is primarily a paracrine mediator of tumor angiogenesis. Thus, anti-VEGFR therapy seems to have a very promising future for the treatment of cancer.
It has been well established that anti-VEGFR2 antibodies suppress tumor angiogenesis by blocking the binding of VEGF to VEGFR23. A variety of drugs that target VEGFR2, such as antibodies and small molecule receptor tyrosine kinase inhibitors, effectively prevent the growth of many mouse tumors and tumor xenografts6,7,8,9. Biochemical analyses have shown that the Ig domain-3 in VEGFR2 (VEGFR2 D3) is important for ligand-binding specificity10. Thus, we chose a recombinant VEGFR2 D3 protein as the target protein. Previously, we successfully isolated an scFv, named AK404R, against VEGFR2 D3. This has a high binding affinity and potent antiangiogenic properties6. However, studies of AK404R have been hampered by its poor pharmacokinetic properties in animal experiments, mainly due to its short half-life. A nanobody is an antibody fragment consisting of a single monomeric variable antibody domain.
As with other recombinant antibody fragments, nanobodies are highly specific because they recognize unique epitopes on target antigens and have the advantages of ease and low cost of manufacturing, lack of immunogenicity and potential for oral dosing formulation. Nanobodies can be used in a broad range of applications for biotechnological and therapeutic purposes: for example, as probes in biosensors, for diagnosing infections and for treating diseases such as cancer or trypanosomiasis11. Therefore, the selection of nanobodies specific for VEGFR2 may provide a novel therapeutic strategy.
Here, we present the first example of VEGFR2-specific nanobodies isolated from the commercial human single domain antibody library HuSdLTM. We demonstrated the ability of one of these nanobodies, named NTV1 (Nanobody Targeting VEGFR), to bind VEGFR2 D3 with high affinity and specificity, as determined by both AlphaScreen and Biacore assays. Furthermore, NTV1 inhibited proliferation and tube formation of human umbilical vein endothelial cell (HUVECs) in vitro. In summary, NTV1 is a potent anti-VEGFR2 D3 nanobody candidate for treating tumors via inhibiting VEGF-mediated angiogenesis.
Materials and methods
Cell lines and expression and purification of soluble VEGFR2 D3
Primary-cultured HUVECs were maintained in Endothelial Cell Medium (ECM, ScienCell Research Laboratories, Carlsbad, CA, US) supplemented with 5% FBS at 37 °C, 5% CO2.
VEGFR2 D3 (amino acid residues 220–330, GenBank accession No AF035121) was cloned into the expression vector pET24a (Novagen) and was designed to be expressed as a His6-SUMO-biotin (for phage display and Biacore assay) or His6-SUMO (for AlphaScreen and functional assays) fusion protein. The recombinant plasmid was transformed into BL21 (DE3) cells. Expressed proteins were obtained as described previously12. Briefly, periplasmic proteins were extracted and loaded on a Nickel HP column (GE Healthcare). After washing with PBS, the bound proteins were eluted with 250 mmol/L imidazole. The eluted fraction was cleaved overnight with a Ulp1 Sumo protease at a protease/protein ratio of 1: 500 in the cold room, loaded on a HiLoad 26/60 Superdex 200 gel filtration column (GE Healthcare) and concentrated using a Qubit® Protein Assay Kit (Life Technologies). The purity of the protein was evaluated using a 15% SDS–PAGE followed by staining with Coomassie Blue R250.
Selection of human anti-VEGFR2 D3 nanobodies from a phage display library
To produce effective antigen-specific recombinant nanobodies with superior binding strength to the antigen VEGFR2 D3, a liquid phase panning strategy, based on a modification of that reported in reference13, was used for all four rounds of panning. Briefly, an affinity matrix (Dynabeads® MyOne™ Streptavidin C1 magnetic beads, Invitrogen) was used for panning. The beads were coated with biotin-VEGFR2 D3 at a concentration of 8 μmol/L in PBS. A large phage display library, the HuSdLTM phage display human single domain antibody library (Creative Biolabs), containing 1×1012 clones was used for selection. After each selection, bound phage particles were immediately used to infect exponentially growing E. coli TG1 cells. The culture was then rescued with an M13KO7 helper phage. It was then amplified, precipitated, and used for the next round of selection. The panning was repeated four times as described above. Enrichment was measured to check the number of phages recaptured after each round of panning, by counting the number of colony forming units (CFU) of the infected TG1 cells. To examine the integrity of the library after the final rounds of panning, 120 individual colonies of E. coli TG1 were randomly isolated. The presence of the nanobody DNA insert in these clones was confirmed by polymerase chain reaction (PCR) amplification and DNA sequencing.
Expression and purification of target nanobodies
Target sequences were cloned into a pETDuet-biotin-MBP expression plasmid, and E. coli BL21 cells were transformed with these plasmids. The fusion proteins were extracted and loaded on an amylose column (GE Healthcare). Further protein purification was performed as described above.
AlphaScreen assay for interactions between VEGFR2 D3 and NTV(1–4)
Interactions between VEGFR2 D3 and NTV(1–4) were assessed by luminescence-based AlphaScreen technology (Perkin Elmer). His6-tagged VEGFR2 D3 was attached to nickel-chelated acceptor beads, and biotinylated NTV(1–4) proteins were attached to streptavidin-coated donor beads. The experiments were conducted with NTV(1–4) proteins and VEGFR2 D3 in the presence of 5 μg/mL donor and acceptor beads. The results were based on an average of three experiments.
Surface plasmon resonance (SPR) kinetic measurements
To determine the binding properties of NTV1, a Biacore experiment was performed using an SPR assay on Biacore T200 (GE Healthcare), according to the manufacturer's instructions. A Biotin CAPture Kit containing CAP chips and reaction solution was obtained from GE Healthcare. Biotin-NTV1 was captured at a concentration of 10 μmol/L at a flow rate of 10 μL/min for 1 min. Meanwhile, one flow cell of the sensor chip was left without captured biotin-NTV1 to provide a reference surface. VEGFR2 D3 was prepared in a series of 7 concentrations using a 3-fold serial dilution. All Biacore kinetic experiment data were obtained using Biacore T200 Evaluation Software to estimate the ka and kd. Data from the three experimental flow cells of a single biosensor chip were globally fit to a 1:1 bimolecular binding model. The KD was calculated from the ratioof kd/ka.
MTT cell proliferation assay
An MTT assay was performed following the method of Mosmann14. Briefly, HUVECs (5×103 cells/well) were plated onto 96-well tissue culture plates (Wallach) in 100 μL 5% FBS-ECM medium and incubated at 37 °C for 24 h. NTV1 at the indicated concentrations or vehicle was added to each well and incubated for an additional 72 h. The color development was measured by reading the absorbance at 570 nm on a micro plate reader (Bio-Rad).
Endothelial tube formation assay (in vitro angiogenesis)
A Geltrix solution (Invitrogen) was thawed on ice, and 50 μL aliqouts were transferred to a 96-well tissue culture plate and incubated at 37 °C for 1 h. For this assay, 4.5×103 HUVECs were added in 100 μL 5% FBS-ECM with NTV1 at indicated concentrations overnight at 37 °C. The following day, endothelial tube formation was digitally photographed with an OLYMPUS inverted microscope. Endothelial cell tube formation was quantified by manual counting of high power fields (×100), and the percentage inhibition is expressed using the untreated group as 100%.
Statistical analysis
All data are expressed as the mean±SEM repeated three times for cell proliferation and endothelial tube formation assays. Statistical analyses were performed using one-way ANOVA followed by a post hoc Dunnett's test. A value of P<0.05 was considered to be statistically significant.
Results
Selection and sequence analysis of VEGFR2 specific nanobodies
To isolate nanobodies against human VEGFR2, we expressed and purified the human VEGFR2 D3 domain (amino acid residues 220–330) as the antigen (Figure 1). After two or three rounds of selection, the antigen-binding clones showed a clear enrichment, as shown in Figure 2. The phage-display library consisted of 1.8×108 clones (CFU/mL) after the fourth round of panning.
Purification and identification of the VEGFR2 D3 proteins with and without biotinylation. The purity of the proteins was determined using Coomassie-stained 15% SDS/PAGE. Size standards are shown on the left side of gel. Lane 1, biotin-VEGFR2 D3 protein; Lane 2, VEGFR2 D3 protein.
Enrichment of phage-display nanobodies by panning. Phage-display nanobodies were selected against biotin-VEGFR2 D3 protein and E coli TG1 was infected with bound phage. The number of phages eluted after each round of panning was counted based on the number of colonies (CFU/mL) formed after reinfection of the host bacteria with the eluted phage particles.
At least eight unique genes (NTV1–8) were identified by DNA sequencing after the final round of panning. Table 1 shows the percentages of these clones based on identical DNA sequences. Of all the positive clones, NTV1 (38/120, 32%) and NTV2 (33/120, 28%) were identified as the most abundant clones. The frequency of NTV5 was 4% (5/120), and NTV(6–8) were each only 2%. Figure 3 shows amino acid sequences deduced from the obtained nucleotide data of NTV(1–8). In the complementarity determining region (CDR), the amino acid sequences had a relatively low sequence homology.
Alignment of amino acid sequences of the NTV(1–4) nanobodies. Amino acid sequences were deduced from the nucleotide sequences. Complementarity determining regions (CDRs) of the variable domain are indicated by white symbols. Framework regions are shown by bold symbols.
Subcloning, expression and purification of NTV(1–4)
For nanobody expression and purification, 4 VEGFR2 D3-blocking clones (NTV1–4) were subcloned into a pETDuet-biotin-MBP expression vector. The recombinant proteins carried an MBP tag to facilitate purification by affinity chromatography. After an additional gel filtration chromatography step, recombinant NTV(1–4) were obtained. No impurity was detected on Coomassie-stained SDS PAGE gels. The recombinant nanobodies were present as a single band of ∼57 kDa (Figure 4). An average yield of approximately 5 mg purified protein was obtained per liter of overnight culture grown in baffle shake flasks.
Purification of NTV(1–4). The purity of the proteins was determined using Coomassie-stained 10% SDS/PAGE.
Affinity measurements
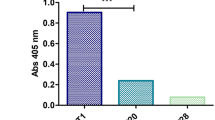
To determine whether the selected nanobodies NTV(1–4) were binders of VEGFR2 D3, we developed an AlphaScreen assay (shown in Figure 5A). As shown in Figure 5B, of the selected nanobody proteins, NTV1 elicited the highest binding signal with VEGFR2 D3 protein at concentrations of either 100 nmol/L or 200 nmol/L. In contrast, NTV1 showed little interaction in the control groups (donor and acceptor beads with VEGFR2 D3 or NTV1 only, or beads without bound protein), indicating specific binding of NTV1 to VEGFR2 D3. NTV2 showed moderate interactions with VEGFR2 D3 (Figure 5C). NTV3 and NTV4 exhibited little or no interaction with VEGFR2 D3 (Figure 5D and 5E).
Interactions between VEGFR2 D3 and NTV(1–4) in the AlphaScreen assay. (A) Cartoon diagram of AlphaScreen assay for detecting NTV(1–4)/VEGFR2 D3 binding. (B–E) NTV(1–4)/ VEGFR2 D3 binding was detected by the AlphaScreen assay. Significant binding signals were detected with biotin-NTV1 and his-tagged VEGFR2 D3 at the indicated concentrations (20, 50, 100 and 200 nmol/L at a ratio of 1:1). The binding assay was repeated three times.
In addition, the SPR assays confirmed the specificity of NTV1/VEGFR2 D3 binding. For this, biotin-NTV1 was immobilized on the CAP sensor chips. The response, in binding units, of NTV1 to VEGFR2 D3 was recorded. NTV1 displayed a high affinity for VEGFR2 D3, yielding an equilibrium dissociation constant (KD) of 49±1.8 nmol/L (Figure 6). By contrast, no binding between VEGFR2 D3 and the biotin control was detected.
Interaction of NTV1 with VEGFR2 D3 by surface plasmon resonance (SPR) analysis. Biotinylated NTV1 was injected at a concentration of 10 μmol/L, and the response level was ∼3500 RU. Sensorgram responses at equilibrium were plotted against VEGFR2 D3 at the indicated concentrations (20, 60, 180, 550, 1600 and 5000 nmol/L). The association and dissociation constant (ka, kd) were calculated using Biacore T200 Evaluation software. KD was calculated from the quotient of kd /ka.
Effects of NTV1 on HUVEC proliferation and endothelial tube formation
VEGFR2 signaling in endothelial cells is primarily responsible for tumor angiogenesis3. The anti-angiogenic effects of NTV1 were determined using HUVECs. As shown in Figure 7, NTV1 inhibited the proliferation of HUVECs.
Effects of NTV1 on HUVEC proliferation. An MTT assay was performed on HUVECs (5×103 cells/well). NTV1 at the indicated concentrations (1, 10, 100 and 1000 nmol/L) or vehicle was added to HUVECs. After 72 h, the intensity of the color was monitored at 570 nm in a micro plate reader. Mean±SEM. n=3. *P<0.05.
As tube formation is one of the late stages of angiogenesis15, we next evaluated the effects of NTV1 on tube formation by HUVECs. In the control group, the HUVECs formed a mesh of tubes within 8 h. Notably, NTV1 at a concentration of 1000 nmol/L almost completely inhibited the formation of the capillary-like structures by the HUVECs (Figure 8).
In vitro anti-angiogenic effects of NTV1 on HUVECs. NTV1 was added to HUVECs (4.5×103 cells/well) at the indicated concentrations (0, 10, 100 and 1000 nmol/L) and incubated overnight. The following day, endothelial tube formation was digitally photographed with an inverted microscope. (A, B) Endothelial cell tube formation was quantified by manual counting in high power fields (×100), (A) 1000 nmol/L NTV1; (B) untreated group: HUVECs without NTV1. (C) Percentage inhibition is expressed using the untreated group as 100%. Mean±SEM. n=3. *P<0.05.
Discussion
The VEGF-VEGFR system plays central roles in pathological angiogenesis in diseases such as cancer. Therefore, a variety of anti-angiogenic drugs, including anti-VEGF/VEGFR antibodies and VEGFR/multi-receptor kinase inhibitors, have been developed and approved for clinical use. Sunitinib, which is a nonselective tyrosine kinase inhibitor that inhibits VEGFR, platelet-derived growth factor receptor and fms-related tyrosine kinase 3 (FLT3), is currently used for the treatment of metastatic renal cancer. Bevacizumab, an anti-VEGFA antibody, is currently used in metastatic colorectal carcinoma (CRC) patients16,17,18,19. While the clinical efficacy of these drugs has been clearly demonstrated in cancer patients, they have not been shown to be effective in curing cancer, suggesting that further improvement in their design is necessary.
In this study, a human anti-VEGFR2 D3 nanobody named NTV1 was isolated from a phage library. This library produces camelized human antibodies that have a human VH3 elongated with HCDR3. Thus, this type of nanobody will gain a competitive advantage over non-human antibodies because of its lower immunogenic potential in humans20, which will be especially useful for long-term and multiple-dose administration. The binding affinity of NTV1 was close to the nanomolar range, thereby approaching the binding affinity of several bivalent anti-VEGFR monoclonal antibodies produced from immunized animals by using hybridoma technology16,17,18,19. This result indicated that the nanobody phage display library can be used to identify functional high-affinity nanobodies that target antigens such as VEGFR.
Considerable clinical and experimental evidence suggests that VEGF-A plays a pivotal role in cancer angiogenesis and metastasis21. Through binding VEGF-A, VEGFR2 is the main signal transducer in tumor-associated angiogenesis. Therapeutic approaches that target the VEGF-A/VEGFR2 signaling axis have been explored for decades. However, efficient and specific small molecule-targeting of this pathway has not yet been achieved22,23. Conventional antibodies against VEGFR2 will block the angiogenic activity of VEGF-A as well as that of other family members exerting their angiogenic effects via the same receptor, including VEGF-C, VEGF-D and VEGF-E18. Gene deletion analysis has demonstrated that the extracellular Ig-like domains 1–3 of VEGFR2 are required for high-affinity binding of VEGF-A, and deletion of Ig domain 3 was found to cause a greater than 1000-fold reduction in binding, indicating the critical role of this domain in VEGFR2/VEGF interaction24. Thus, we used VEGFR2 D3 as the binding target to obtain a nanobody that might have binding affinity for VEGF-A with high specificity. Our results demonstrated that NTV1 bound to VEGFR2 D3 with high affinity, indicating that NTV1 might exert its VEGF-A blocking activity by steric hindrance resulting from the interaction between the antibody and the receptor. This hypothesis is worthy of further confirmation by structural analysis of the NTV1/VEGFR2 D3 complex.
VEGFR2-neutralizing antibodies have shown encouraging inhibitory effects on solid tumor growth, ascites formation and metastatic dissemination in several animal models7,8,25. However, the results in human cancer have been less impressive20. One possible explanation might be the large size of conventional antibodies, which are unlikely to penetrate the vascular barrier efficiently26. Additional limitations of monoclonal antibodies against VEGFR2 are their serious (although uncommon) side effects and high costs27. Nanobodies have many advantageous properties, such as a low molecular mass (<60 kDa), strict monomeric behavior, low immunogenicity, high affinity, high solubility and stability and high yield in bacteria or yeast28,29,30. Here, we demonstrated that NTV1 effectively inhibited the proliferation and tube formation of HUVECs. The anti-angiogenic effects of NTV1 suggest that NTV1 is a functional nanobody candidate for blocking tumor angiogenesis.
In conclusion, we described the successful isolation of the VEGFR2 D3-specific nanobody NTV1 from a human single domain antibody library. Following the expression of soluble NTV1 protein, the characterization of its binding to VEGFR2 D3, by AlphaScreen and surface plasmon resonance, and its function, by proliferation and tube formation inhibition, were carefully assessed. Taken together, the results show that NTV1 is a potential therapeutic candidate for blocking VEGFR2 D3, thereby providing a novel strategy for the development of VEGFR2-targeted nanobody-based cancer therapeutics. Further studies focusing on improving the binding affinity and specificity of this nanobody by multiple methods, such as gene evolution, are ongoing. The current study provides a basis for further improvements in functionality and stability. Structural analysis of the NTV1/VEGFR2 D3 complex and clinical evaluation of NTV1 will also be worthy of further investigation.
Author contribution
Lin MA, Kai GU, Xue-tao CHEN, Yi JIANG: substantial contributions to conception and design, or acquisition of data, or analysis and interpretation of data; Cheng-hai ZHANG, Karsten MELCHER, Juan ZHANG, Min WANG: involvement in drafting the article or revising it critically for important intellectual content; Lin MA and H Eric XU: final approval of the version to be published.
References
Yang H, Shin S, Ahn J, Choi Y, Kim KH, Chung CJ . Local injection of pulp cells enhances wound healing during the initial proliferative phase through the stimulation of host angiogenesis. J Endod 2013; 39: 788–94.
Shaik-Dasthagirisaheb YB, Varvara G, Murmura G, Saggini A, Potalivo G, Caraffa A, et al. Vascular endothelial growth factor (VEGF), mast cells and inflammation. Int J Immunopathol Pharmacol 2013; 26: 327–35.
Kopparapu PK, Boorjian SA, Robinson BD, Downes M, Gudas LJ, Mongan NP, et al. Expression of VEGF and its receptors VEGFR1/VEGFR2 is associated with invasiveness of bladder cancer. Anticancer Res 2013; 33: 2381–90.
Honda S, Nagai T, Negi A . Anti-angiogenic effects of non-peptide integrin alphavbeta3 specific antagonist on laser-induced choroidal neovascularization in mice. Graefes Arch Clin Exp Ophthalmol 2009; 247: 515–22.
Clavel G, Bessis N, Lemeiter D, Fardellone P, Mejjad O, Menard JF, et al. Angiogenesis markers (VEGF, soluble receptor of VEGF and angiopoietin-1) in very early arthritis and their association with inflammation and joint destruction. Clin Immunol 2007; 124: 158–64.
Zhang J, Li H, Wang X, Qi H, Miao X, Zhang T, et al. Phage-derived fully human antibody scFv fragment directed against human vascular endothelial growth factor receptor 2 blocked its interaction with VEGF. Biotechnol Prog 2012; 28: 981–9.
Gule MK, Chen Y, Sano D, Frederick MJ, Zhou G, Zhao M, et al. Targeted therapy of VEGFR2 and EGFR significantly inhibits growth of anaplastic thyroid cancer in an orthotopic murine model. Clin Cancer Res 2011; 17: 2281–91.
Lynn KD, Udugamasooriya DG, Roland CL, Castrillon DH, Kodadek TJ, Brekken RA . GU81, a VEGFR2 antagonist peptoid, enhances the anti-tumor activity of doxorubicin in the murine MMTV-PyMT transgenic model of breast cancer. BMC Cancer 2010; 10: 397.
Zhang W, Ran S, Sambade M, Huang X, Thorpe PE . A monoclonal antibody that blocks VEGF binding to VEGFR2 (KDR/Flk-1) inhibits vascular expression of Flk-1 and tumor growth in an orthotopic human breast cancer model. Angiogenesis 2002; 5: 35–44.
Ge YL, Zhang JY, Zhang X, Hou L, Li Q, Xue ML . Chemically modified siRNA directed against the KDR gene inhibits the proliferation of breast cancer cells. Mol Med Rep 2009; 2: 121–7.
Tillib SV, Ivanova TI, Vasilev LA . Fingerprint-like analysis of “Nanoantibody” selection by phage display using two helper phage variants. Acta Naturae 2010; 2: 85–93.
Ban T, Ke J, Chen R, Gu X, Tan MH, Zhou XE, et al. Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J Biol Chem 2013; 288: 31540–8.
Dennis MS, Lazarus RA . Kunitz domain inhibitors of tissue factor-factor VIIa. I. Potent inhibitors selected from libraries by phage display. J Biol Chem 1994; 269: 22129–36.
Mosmann T . Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65: 55–63.
Watari K, Nakamura M, Fukunaga Y, Furuno A, Shibata T, Kawahara A, et al. The antitumor effect of a novel angiogenesis inhibitor (an octahydronaphthalene derivative) targeting both VEGF receptor and NF-kappaB pathway. Int J Cancer 2012; 131: 310–21.
Choi HJ, Kim YJ, Lee S, Kim YS . A heterodimeric Fc-based bispecific antibody simultaneously targeting VEGFR-2 and Met exhibits potent antitumor activity. Mol Cancer Ther 2013; 12: 2748–59.
Huang J, Tan Y, Tang Q, Liu X, Guan X, Feng Z, et al. A high-affinity human/mouse cross-reactive monoclonal antibody, specific for VEGFR-2 linear and conformational epitopes. Cytotechnology 2010; 62: 61–71.
Klement G, Huang P, Mayer B, Green SK, Man S, Bohlen P, et al. Differences in therapeutic indexes of combination metronomic chemotherapy and an anti-VEGFR-2 antibody in multidrug-resistant human breast cancer xenografts. Clin Cancer Res 2002; 8: 221–32.
Lambertini C, Barzaghi-Rinaudo P, D'Amato L, Schulz S, Nuciforo P, Schmid HA . Evaluation of somatostatin receptor subtype expression in human neuroendocrine tumors using two sets of new monoclonal antibodies. Regul Pept 2013; 187: 35–41.
Zhu AX, Finn RS, Mulcahy M, Gurtler J, Sun W, Schwartz JD, et al. A phase II and biomarker study of ramucirumab, a human monoclonal antibody targeting the VEGF receptor-2, as first-line monotherapy in patients with advanced hepatocellular cancer. Clin Cancer Res 2013; 19: 6614–23.
Srabovic N, Mujagic Z, Mujanovic-Mustedanagic J, Softic A, Muminovic Z, Rifatbegovic A, et al. Vascular endothelial growth factor receptor-1 expression in breast cancer and its correlation to vascular endothelial growth factor a. Int J Breast Cancer 2013; 2013: 746749.
Sitohy B, Nagy JA, Dvorak HF . Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res 2012; 72: 1909–14.
Sharma PS, Sharma R, Tyagi T . VEGF/VEGFR pathway inhibitors as anti-angiogenic agents: present and future. Curr Cancer Drug Targets 2011; 11: 624–53.
Lu D, Kussie P, Pytowski B, Persaud K, Bohlen P, Witte L, et al. Identification of the residues in the extracellular region of KDR important for interaction with vascular endothelial growth factor and neutralizing anti-KDR antibodies. J Biol Chem 2000; 275: 14321–30.
Orecchia P, Conte R, Balza E, Petretto A, Mauri P, Mingari MC, et al. A novel human anti-syndecan-1 antibody inhibits vascular maturation and tumour growth in melanoma. Eur J Cancer 2013; 49: 2022–33.
Kim JH, Kim JH, Kim DH, Park WY, Kim KW, Yu YS . Intravenously administered anti-recoverin antibody alone does not pass through the blood-retinal barrier. Korean J Ophthalmol 2011; 25: 189–95.
Dienstmann R, Brana I, Rodon J, Tabernero J . Toxicity as a biomarker of efficacy of molecular targeted therapies: focus on EGFR and VEGF inhibiting anticancer drugs. Oncologist 2011; 16: 1729–40.
Oliveira S, Heukers R, Sornkom J, Kok RJ, van Bergen En Henegouwen PM . Targeting tumors with nanobodies for cancer imaging and therapy. J Control Release 2013; 172: 607–17.
Vosjan MJ, Vercammen J, Kolkman JA, Stigter-van Walsum M, Revets H, van Dongen GA . Nanobodies targeting the hepatocyte growth factor: potential new drugs for molecular cancer therapy. Mol Cancer Ther 2012; 11: 1017–25.
Cortez-Retamozo V, Backmann N, Senter PD, Wernery U, De Baetselier P, Muyldermans S, et al. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res 2004; 64: 2853–7.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (81072561, 81102364, 31300607 and 31230022).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ma, L., Gu, K., Zhang, Ch. et al. Generation and characterization of a human nanobody against VEGFR-2. Acta Pharmacol Sin 37, 857–864 (2016). https://doi.org/10.1038/aps.2016.2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2016.2