Abstract
Aim:
To investigate the effects of serum deprivation (SD) on microvesicles (MVs) secreted from human myeloma cells and the implications for disease progression.
Methods:
RPMI 8226, U266, and KM3 human myeloma cells were incubated in medium containing 10% (non-SD) or 1% fetal bovine serum (SD) and MVs were isolated. The levels and size distribution of MVs were analyzed with flow cytometry. The protein profiles of MVs were studied using 2D SDS-PAGE, MALDI-TOF-MS, and Western blotting. NF-κB activation was analyzed using EMSA. Angiogenesis was examined in Eahy926 endothelial cells.
Results:
Exposure of RPMI 8226 cells to SD for 24 h did not alter the number of apoptotic cells. However, SD increased the number of MVs from RPMI 8226, U266, and KM3 cells to 2.5-, 4.3-, and 3.8-fold, respectively. The size distribution of SD MVs was also significantly different from that of non-SD MVs. Three proteins ZNF224, SARM, and COBL in SD MVs were found to be up-regulated, which were involved in cell cycle regulation, signal transduction and metabolism, respectively. Co-culture of SD MVs and RPMI 8226 cells increased NF-κB activation in the target RPMI 8226 cells. Furthermore, SD MVs from RPMI 8226 cells significantly increased the microtubule formation capacity of Eahy926 endothelial cells compared with non-SD MVs.
Conclusion:
SD elevates the levels of microvesicles with different size distribution and selectively enriched proteins in human myeloma cells in vitro. The selectively enriched proteins, especially ZNF224, may play key roles in regulation of myeloma cells, allowing better adaptation to SD.
Similar content being viewed by others
Introduction
One hallmark of human cancers is their ability to evade apoptosis1. Cancer cells adapt better than do normal diploid cells to environmental stress. When confronted with pro-cell death signals, cancer cells initiate complicated anti-apoptotic processes; promote angiogenesis; and alter their energy metabolism to survive, invade or even metastasize2. Multiple myeloma (MM) is a plasma cell malignancy that occurs in the bone marrow, and conventional high-dose chemotherapy ultimately results in drug resistance3. Studies have shown that a decrease in apoptosis may be an indicator of progression from the inactive disease monoclonal gammopathy of undetermined significance (MGUS) to overt MM4. In other words, certain defects in the apoptotic process could be involved in this neoplasia.
In the in vitro cultivation of most cell lines and primary cells, fetal bovine serum (FBS) is frequently used as supplementation to a basal medium. FBS contains certain nutritional and macromolecular growth factors that facilitate cell growth. Serum deprivation (SD), which has also been referred to as environmental stress, is a common model of cellular stress in vitro and is used to study the acquired anti-apoptosis of tumor cells5,6. Cellular stress has been identified as one of the key factors responsible for initiating the carcinogenic process and treatment resistance1. SD reduces basal cellular activity and induces apoptosis in various cells7. While in the early stage of SD, in contrast to normal cells, tumor cells initiate complicated processes against apoptosis2,8.
Microvesicles (MVs), also known as shedding vesicles or microparticles, are endosome-derived organelles that are spontaneously secreted from normal and neoplastic cells. These vesicles were originally considered inert cellular debris; however, it is now known that both their cytoplasmic fragments and their membranes contain a variable spectrum of molecules whose pattern specifically reflects the donor cell that secretes these MVs9. Release of MVs is a process that occurs in a spectrum of cell types and, more frequently, in tumor cells. Recently, rapidly emerging evidence has supported the significance of MVs in a broad range of physiological and pathological processes10. Important work has been devoted to the biology of tumor cell-derived MVs. Compelling evidence suggests that tumor cell-derived MVs have vital roles in tumor invasion and metastasis, and their pathological functions are still under intense investigation11. However, it is not yet known how MM cell-derived MVs change under SD and whether these MVs are responsible for tumor progression and treatment resistance.
In this study, using well-documented myeloma cell lines as cell models, we determined whether and how SD affects the characteristics of myeloma cell-derived MVs, including their levels, size distribution and enriched proteins. In this context, we evaluated the role of ZNF224 in MVs and how this protein affects NF-κB activity in target myeloma cells and tube formation in endothelial cells.
Materials and methods
Cell lines and culture
RPMI 8226 and U266 human myeloma cell lines were obtained from ATCC (USA), and KM3 myeloma cells were a kind gift from Dr Jian HOU (Department of Hematology, The Myeloma & Lymphoma Center, Changzheng Hospital, Shanghai, China). The cells were maintained in RPMI-1640 medium supplemented with 10% FBS (Gibco, Carlsbad, CA, USA) and 1% penicillin-streptomycin (Beyotime, Shanghai, China) at 37 °C in a humidified atmosphere in the presence of 5% CO2. The cells were passaged three times per week. SD was performed under the same culture conditions but with 1% FBS.
Proliferation, apoptosis and cell cycle assays
To assay the effect of SD on RPMI 8226 cells, cell proliferation was evaluated by an MTT assay, and the cell cycle and apoptosis were analyzed by flow cytometry. In the MTT assay, RPMI 8226 cells were seeded at 5×104 cells/well into a 96-well tissue culture plate. MTT reagent (Sigma-Aldrich, St Louis, MO, USA) was added 24 h later and the culture was maintained for 4 h. The formazan was dissolved in DMSO and the optical density was assessed at a wavelength of 570 nm. For apoptosis analysis, RPMI 8226 cells were washed twice in PBS, and the Annexin V-FITC apoptosis detection kit (KeyGEN, Nanjing, China) was used according to the manufacturer's instructions. The percentage of cells positive for Annexin V was analyzed by flow cytometry. Cell cycle analysis was performed with propidium iodide (PI) staining (50 μg/mL) to assess the percentage of cells at each cell cycle stage. For each sampling, 1×104 cells were analyzed using a Becton Dickinson FACSCalibur (Mountain View, CA, USA).
Isolation of MVs
MVs were isolated from the conditioned medium of myeloma cells by continuous differential centrifugation, as described9. Briefly, myeloma cells harvested in the log growth phase were reseeded at 5×105 cells/mL in RPMI-1640 with 10% FBS or 1% FBS (we also removed FBS-derived MVs by the method of differential centrifugation before use). After 24 h of culture, cells and debris were removed by centrifugation at 750×g for 15 min and 1500×g for 20 min, respectively. MVs were pelleted by further centrifugation at 16 000×g for 45 min at 4 °C. Trypan blue was used for dead cell counting after SD. Only samples with <5% cell death were used, which ensured that our MVs were predominantly caused by stimulation and not cell death.
Transmission electron microscopy
To compare the microscopic characteristics of SD MVs and non-SD MVs (control), RPMI 8226 cells maintained in medium containing 10% or 1% FBS were fixed in 3% glutaraldehyde for 2 h and then in 1% osmium tetroxide. Following serial dehydration, the cells were soaked in propylene oxide and then embedded in resin. Sections of 0.1 μm in thickness were prepared from the cells. Shedding of MVs was observed around the cell membrane. Photographs (×25 600) were taken using a Fei.Tecnai.GZ.12 transmission electron microscope (FEI, Hillsboro, OR, USA).
Flow cytometry analysis of MVs
Standard microbeads with a diameter of 1 μm (Sigma-Aldrich, St Louis, MO, USA) were used to set the upper size limit for MVs. To determine the number of MVs, aliquots (5 μL) of 3 μm standard microbeads (6.77×109 particles/mL; Sigma-Aldrich, St Louis, MO, USA) were added to an MV sample. MVs isolated from a 10 mL supernatant of myeloma cell-conditioned medium were resuspended in PBS and then stained with calcein-AM (5 μg/mL; Life Technologies, Carlsbad, CA, USA) for 20 min at ambient temperature. The stained MVs were diluted in 400 μL PBS and analyzed using a FACSAria II (BD Pharmingen, San Diego, CA, USA). MVs were defined as calcein-positive events. The counting endpoint was set as 100 000 events in the population corresponding to the 3 μm standard beads. The relative number in the two samples was calculated using the 3 μm beads as an internal standard12. The concentration of MVs in a sample can be calculated with the formula below:

Cs: Concentration of standard microbeads in the sample to be analyzed; here, we prepared each sample with a Cs of 8.46×107 particles/mL. Cmv: Concentration of MV events in the sample to be analyzed. Eventp2: Event number in the P2 population (3 μm standard microbeads); here, we set a counting endpoint of 100 000 events for the P2 population. Caleventp1: Calcein-AM-positive event number in the P1 population.
For size assessment, Dragon Green-coupled polystyrene microbeads (0.2, 0.5, and 0.8 μm; Bangs Laboratories, Fishers, IN, USA) were used for size calibration. Gating and analysis were determined by a reported method9. Flow cytometry data were analyzed using FlowJo software, version 7.6 (Tree Star Inc, Ashland, OR, USA).
Two-dimensional polyacrylamide gel electrophoresis
MVs produced by RPMI 8226 cells in culture medium with 10% FBS or 1% FBS were pelleted by ultracentrifugation, as described above. The resulting pellet was lysed in lysis buffer (4% CHAPS, 4 mol/L Tris, 8 mol/L urea and 1% DTT), and the lysates were centrifuged at 12 000×g for 30 min at 4 °C. The supernatant was transferred to a new tube, and the total protein concentration was determined by Bradford assay. Triplicate gels were made for each MV sample, and a total of 150 μg protein per gel was analyzed. Linear IPG 18-cm strips (pH 3–10) were rehydrated with MV protein samples for 12 h at 30 V. The gel was run according to the manufacturer's instructions on an IPGphor II apparatus (GE Healthcare, fairfield, CT, USA) in gradient mode: 200 V step-n-hold for 1 h, 500 V gradient for 1 h, 1500 V gradient for 0.5 h, 8000 V gradient for 1 h and 8000 V step-n-hold for 7 h. Focused IPG strips were equilibrated for 15 min in a solution [6 mol/L urea, 2 % SDS, 30% glycerol, 50 mmol/L Tris-HCl (pH 8.8) and 0.2 % DTT] and then for an additional 15 min in the same solution, except that DTT was replaced by 3% iodoacetamide. Two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was then performed using the Ettan DALTsix electrophoresis system (GE Healthcare, fairfield, CT, USA) in a Hoefer DALT™ tank. To visualize the protein spots in the 2-DE gels, the gels were silver stained.
Gel scanning and image analysis
The stained 2-DE gels were scanned and analyzed using the PDQuest system (Bio-Rad,Hercules, CA, USA), following the manufacturer's protocols. Three separate gels were prepared for each MV sample to minimize the contribution of experimental variation. The gel spot pattern of each gel was summarized in a standard after spot matching. The relative intensities were quantified as the intensity of a spot relative to the total intensity of all the spots in the gel. The change index was defined as the ratio of the relative intensities of the spots from SD MVs and non-SD MVs. All indexes were calculated based on the mean of the spot intensities of all six gels, with standard deviations. Proteins were classified as being differentially expressed when the spot intensity showed ≥2-fold variation between SD MVs and control MVs.
Mass spectrometric determination of partial peptide sequence
Protein spots showing comparable differences across SD MVs and non-SD MVs were excised from the gels, and in-gel digestion was performed. The gel spots were washed three times with double-distilled water and destained with a fresh solution containing 100 mmol/L ferricyanide kalium and 50 mmol/L sodium hyposulfite. The pieces were then washed twice in 25 mmol/L sodium bicarbonate and 50% methyl cyanides. After being dried in a vacuum centrifuge, the gels were digested overnight with gentle agitation in 20 mg/L trypsin (Promega, Madison, WI, USA) at 37 °C. The tryptic peptide mixture was extracted three times with 0.1% trifluoroacetic acid in 50% acetonitrile and then dried overnight. The dried material was dissolved in a saturated solution of a-cyano-4-hydroxycinnamic acid with 0.1% trifluoroacetic acid and 50% acetonitrile. Peptide mass fingerprinting (PMF) was performing using a Bruker Daltonics Autoflex TOF-TOF LIFT MS (Bruker, Karlsruhe, BW, Germany). For database searching, the MASCOT search engine was used by setting a peptide tolerance of 50 ppm.
Western blot analysis
Myeloma cells or MVs were solubilized in SDS lysis buffer and quantified by Bradford assay. Lysed protein samples (40 μg) were subjected to 10% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked for 1 h at room temperature in TBST [50 mmol/L Tris-HCl (pH 7.6), 150 mmol/L NaCl and 0.1% Tween 20] and 5% nonfat dried milk and then incubated with rabbit polyclonal anti-human ZNF224 antibody (Sigma-Aldrich, St Louis, MO, USA) (1:400 in blocking buffer overnight at 4 °C), followed by rabbit polyclonal anti-human β-actin antibody (Santa Cruz Biotechnologm, Santa Cruz, CA, USA) (1:1000 in blocking buffer overnight at 4 °C). The membranes were incubated with the appropriate secondary antibody for 2 h at room temperature, followed by enhanced chemiluminescence according to the manufacturer's instructions (Beyotime, Shanghai, China). The densities of the immunoreactive bands were measured by PDQuest software (Bio-Rad,Hercules, CA, USA).
Electrophoretic mobility shift assays
After co-culture with 100 μg/mL SD MVs or non-SD MVs for 12 h, nuclear extracted proteins from RPMI 8226 cells were prepared using a nuclear protein extraction kit (Beyotime, Shanghai, China) and quantified by Bradford assay. Co-culture of the same concentration of heat-inactivated (HI)-non-SD MVs or HI-SD MVs was set as a control. Heat inactivation of MVs was performed at 100 °C for 10 min13. Electrophoretic mobility shift assays (EMSAs) were performed using a LightShift kit (Pierce, Rockford, IL, USA). In particular, 5 μg samples were incubated in 20 μL of reaction mixture with or without an excess of DNA competitors on ice for 20 min. Biotin-labeled probes (synthesized and 5′-labeled by Sangon Inc, Shanghai, China) were then added. For super-shift assays, 5 μg of antibodies was added 10 min before the addition of the probe to the reaction mixture. All DNA–protein complexes were resolved by electrophoresis on 5% native polyacrylamide and transferred to Immobilon-Ny+ membranes (Millipore, Billerica, MA, USA).
Quantitative real-time polymerase chain reaction
The total RNA of RPMI 8226 cells cocultured with 100 μg/mL SD MVs or non-SD MVs for 12 h or 24 h was isolated with TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. First-strand cDNA synthesis was performed using the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan). Real-time polymerase chain reaction (RT-PCR) assays were performed using SYBR Premix Ex TaqTM (Takara, Otsu, Japan) on a real-time PCR system (ABI 7500, Applied Biosystems, USA). The primers (Invitrogen, Carlsbad, CA, USA) used were as follows: VEGF, 5′-AAG GAG GAG GGC AGA ATC AT-3′ (F) and 5′-ATC TGC ATG GTG ATG TTG GA-3′ (R); IL-6, 5′-ATG AGG AGA CTT GCC TGG TG-3′ (F) and 5′-CAG GGG TGG TTA TTG CAT CT-3′ (R); and GAPDH, 5′-AGA AGG CTG GGG CTC ATT TG-3′ (F) and 5′-AGG GGC CAT CCA CAG TCT TC-3′ (R). The real-time PCR conditions were 2 min at 50 °C, 10 min at 95 °C and then 40 cycles of 30 s at 95 °C and 30 s at 60 °C. Quantitation was performed using the comparative CT method, with GAPDH as the normalization gene. Log2-transformed fold-change values were plotted and used for significant differences tests. Additionally, mRNA expression was confirmed by semiquantitative RT-PCR.
Tube formation assays
Matrigel-coated plates were prepared by adding 100 μL/well Matrigel (BD, USA) to 96-well plates. Aliquots of Eahy926 cells (1×104) were seeded onto the Matrigel-coated wells of a 96-well plate and cultured in 10% FBS-supplemented DMEM in the presence of 100 μg/mL non-SD MVs or SD MVs. The same concentration of HI-non-SD MVs and HI-SD MVs was set as a control. Triplicate wells were used in each experiment. Tube formation was examined after 6 h of culture by phase-contrast microscopy (Olympus IX81, Tokyo, Japan), and closed tube formation was quantified.
Statistical analysis
The data are expressed as mean±SEM of at least three experiments. Statistical analyses were performed using a paired Student's t test, and differences were considered statistically significant at P<0.05.
Results
SD treatment results in significantly decreased proliferation and G0/G1 state arrest without increased apoptosis in RPMI 8226 cells
To confirm the effect of SD on RPMI 8226 cells, we exposed RPMI 8226 cells to SD (1% FBS) for 24 h and tested the influence of SD on cell proliferation, the cell cycle and apoptosis. In the MTT assay, RPMI 8226 cells that underwent 24 h of SD showed 89% of the cell proliferative response in non-SD cultures (P<0.05) (Figure 1A), suggesting that SD significantly reduced the proliferation of myeloma cells. Shortages of nutritional support and macromolecular growth factors due to a lack of FBS may have been the cause of the low proliferation rate14.
SD treatment resulted in significantly decreased proliferation and G0/G1 state arrest without increased apoptosis in RPMI 8226 cells. RPMI 8226 cells were cultured with 10% FBS and 1% FBS (SD) for 24 h, and proliferation and apoptosis were analyzed using MTT and flow cytometry, respectively. (A) For the growth assays, MTT was used and the results were indicated. (B) Cell cycle was determined by FACS analysis of DNA fragmentation of propidium iodide-stained nuclei. SD resulted in increased RPMI 8226 cells at the G0/G1 phase (50.75%±1.13% vs 62.23%±1.26%, P<0.01) and increased cells at the G2/M phases (18.24%±0.65% vs 9.43%±0.40%, P<0.01). (C and D) For apoptosis analysis, the cells under SD and non-SD were stained with annexin V and evaluated using FACS. No induced apoptosis of RPMI 8226 cells under SD was observed (5.56%±0.83% vs 6.80%±0.16%, P>0.05). The data shown represent the mean±SEM from at least 3 independent experiments. aP>0.05, bP<0.05, Student's t-test.
Additionally, we observed that 24 h of SD caused an increase in the number of RPMI 8226 cells in the G0/G1 phase (50.75%±1.13% vs 62.23%±1.26%) (P<0.01) and a decrease in the number of cells in the S and G2/M phases (18.24%±0.65% vs 9.43%±0.40%) (P<0.01) (Figure 1B), which is consistent with the current standard approach to the synchronization and arrest of cells in the G0/G1 state through SD treatment15.
We observed no significant increase in the number of annexin V-positive cells (5.56%±0.83% vs 6.80%±0.16%) (P>0.05) after 24 h of SD (Figure 1C and 1D). Our hypothesis is that the changes in myeloma cell-derived MVs in the early stage of SD may reflect the adapted phenotype of the myeloma cells, which could be targeted by therapy. Because the pro-apoptotic effect of long hours of SD was obvious for RPMI 8226 cells16, 24 h was chosen as the time point for MV collection. Dead cells were kept below 5% for both groups, which ensured that SD MVs were predominantly caused by stimulation and not cell death.
Validation of MVs
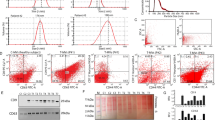
Both the non-SD MVs and the SD MVs from RPMI 8226 cells showed a spheroid shape with a diameter of 0.1–1.0 μm in transmission electron microscopy images (Figure 2A and 2B). The MVs were located close to the cell membrane, which is consistent with other reports17. Using 1 μm beads as the internal size standard, we analyzed MVs below the FSC signal, corresponding to 1 μm beads (P1 population in Figure 2E). Furthermore, calcein-AM was used to define MVs as calcein-AM-positive events. Calcein-AM is a non-fluorescent calcein acetoxymethyl ester that is membrane permeable and has negligible membrane binding. Upon cleavage by cytosolic esterases, calcein binds to calcium, becomes fluorescent, and is retained within MVs. Thus, only intact MVs will fluoresce, and concomitant staining of debris is avoided9. The data suggested that our MV sample collected following the differential centrifugation protocol had a purity of 70% (Figure 2F), and cell debris or mass-molecule compounds were observed as calcein-negative events in the FACS analysis.
Characteristics of MVs generated by myeloma cells under SD and non-SD. The transmission electron microscopy images of MVs generated by RPMI 8226 cells under SD and non-SD are respectively shown in (A) and (B). Flow cytometry plots of three standard microbeads (0.2, 0.5, and 0.8 μm) and distribution of the three standard microbeads on the histogram in flow cytometry analysis are respectively shown in (C and D). (E) Representative flow cytometry plots of MVs. The microbeads (1 μm) were used as a size internal standard (P1 proportion) and equal count of the microbeads (3 μm) (P2 proportion) were used as an internal standards. (F) Integrity validation of MVs sample by calcein-AM. MVs were defined as calcein-positive events. Only intact MVs is calcein-AM positive and concomitant staining of debris was avoided. Counting end was set as 105 events in the poportion of the 3 μm standard beads. The data suggested that the samples, including SD MVs and non-SD MVs derived from myeloma cells (RPMI 8226, U266, and KM3), acquired by differential centrifugation method had 70% of intact MVs. (G) Equal count of 3 μm beads (P2 proportion) was used as a counting internal standards, MVs events were counted for both samples and the corresponding bars was shown. SD stimulated more MVs than non-SD in myeloma cells (RPMI 8226: 2.5-fold; U266: 4.3-fold; KM3: 3.8-fold). Data represent mean±SEM. n=3. cP<0.01.
SD induced elevated levels of MVs with different size distributions from RPMI 8226, U266, and KM3 myeloma cells
Three sizes of standard microbeads (0.2, 0.5, and 0.8 μm; Bangs Laboratories, Fishers, IN, USA) were used for size calibration (Figure 2C). When we analyzed the size distribution of a mixed sample of the three standard microbeads, three distinct distributions of microbeads were observed in the histogram (Figure 2D), validating our method of testing the size distribution of MVs. Using equal counts of larger (3 μm) beads as an internal standard (P2 population in Figure 2E), we analyzed the number of calcein-AM-positive events recorded in the supernatants of myeloma cells by flow cytometry. The data suggested that SD stimulated 2.5-fold more MVs from RPMI 8226 cells than did the control (P<0.01) and 4.3- and 3.8-fold more MVs from U266 and KM3 cells, respectively, than did the control (Figure 2G). Thus, SD stimulated RPMI 8226 cells to secrete significantly higher levels of MVs.
Furthermore, the data suggested that all three myeloma cell lines generated distinct populations of MVs with different size distributions when confronted with SD (Figure 3). This study found that cell stress affected the size distribution of cell-derived MVs and evidenced that myeloma cells confronted with SD shed different-sized MVs.
Distinct size distribution of MVs generated by myeloma cells under SD and non-SD. (A) Non-SD MVs, (B) SD MVs, (C) Corresponding bar charts of the different size distribution of non-SD MVs and SD MVs. Data represent mean±SEM. n=3. bP<0.05, cP<0.01.
Selectively enriched proteins in SD MVs
Because SD resulted in elevated levels of MVs with different size distributions from myeloma cells, we used RPMI 8226 cells to determine the proteins in SD MVs and to understand the functional implication of SD-induced MVs for myeloma. We hypothesized that different proteins in the MVs from RPMI 8226 cells were induced by SD. To identify the proteins involved, we initiated a comparative proteomic study of MVs derived from RPMI 8226 cells that did or did not undergo SD. Two representative 2-DE maps from the two groups are shown in Figure 4A. For a reliable analysis of protein expression, 2-DE was performed for each group in triplicate. The attained 2-DE patterns of each group were highly reproducible and well resolved. Close-up views of the region of the gels showing differential expression of proteins between the two MV populations are shown in Figure 4B. Using PDQuest software, version 8.0.1, the expression level of each protein was normalized as the percentage of the spot's volume relative to the total volume of valid protein spots in the gel (Figure 4C). Image analysis revealed that five spots (spots 1, 2, 3, 4, and 5) had specifically increased in size (≥2-fold upregulated, P<0.05) for SD MVs, whereas two spots (spots 6 and 7) had decreased in size (≥2-fold downregulated, P<0.05) compared with spots for non-SD MVs. The results suggested that selected proteins were expressed in the SD MVs.
Comparative proteomic study of MVs. (A) Representative 2-DE gel protein expression maps of the MVs generated under SD and non-SD condition. The arrows indicate the positions of all 7 protein spots that were significantly altered under SD. (B) Close view in the representative 2-DE gels of differentially expressed proteins derived from the two populations of MVs. (C) Corresponding bar charts of relative expression levels of the 7 protein spots. The data represents the mean±SEM from three independent experiments. bP<0.05.
The seven differentially expressed protein spots, marked with numbers and arrows in Figure 4A, were excised from silver-stained gels, in situ digested with trypsin and analyzed by matrix-assisted laser desorption ionization-time-of-flight-mass spectrometry (MALDI-TOF-MS). High-quality PMF was performed for three (1, 2, and 3) of the differentially expressed proteins. The MALDI-TOF-MS maps of spots 1, 2, and 3 are shown in Figure 5A, 5B, and 5C, respectively. The monoisotopic peaks were input into the Mascot search engine to search the Swiss-Prot database, and the query results showed that the three up-regulated proteins in SD MVs were ZNF224, SARM and COBL. The annotation of the three identified proteins is summarized in Table 1. Based on protein functions determined using information from the Swiss-Prot database, the identified proteins associated with a response to SD were involved in cell cycle regulation, signal transduction, and metabolism18,19,20, which suggested that the enrichment of these proteins in SD MVs was involved in myeloma progression.
Matrix-assisted laser desorption-time of flight-mass spectrometry (MALDI-TOF-MS) analysis of differentially expressed proteins. (A), (B), and (C) showed MALDI-TOF-MS mass spectrum of spot 1, 2, and 3, respectively.
Validation of enriched ZNF224 protein in SD MVs
Among the three proteins identified above, ZNF224 is a known transcriptional repressor that was reported to have a positive effect on the activity of the NF-κB signaling pathway21. Here, considering the vital role of NF-κB activation in MM disease advancement and the transcriptional activation of anti-apoptotic proteins by NF-κB22,23, we performed a Western blot analysis of ZNF224 to further validate this protein's presence in SD MVs. The results confirmed ZNF224 as an upregulated protein in myeloma cell-derived MVs after SD (Figure 6). Interestingly, we also observed up-regulation of ZNF224 in the parent cells after SD (Figure 6). When comparing ZNF224 protein expression levels relative to β-actin levels, we found that the expression of ZNF224 in non-SD MVs was at the same level as in the parent cells; however, the level was 3-fold higher in SD MVs than that in the parent RPMI 8226 and U266 cells (1 and 2 in Figure 6). In other words, myeloma cells selectively and significantly excreted the ZNF224 protein via shedding MVs when confronted with SD, suggesting that it was of great importance to investigate this selective expression of the ZNF224 protein.
Validation and evaluation of ZNF224 protein of MVs and myeloma cells. (A) Protein extracts were obtained from myeloma cells and MVs from myeloma cells under SD and non-SD for 24 h, respectively. ZNF224 protein expression levels were studied by Western blotting, with β-actin as control. (B) Normalized levels of ZNF224 expression with β-actin. The bands were analyzed using PDquest software. Data represents the mean±SEM from three independent experiments performed in triplicate. cP<0.01.
Co-culture of SD MVs increased NF-κB activation in RPMI 8226 cells
Because ZNF224 was reported to positively regulate the NF-κB signaling pathway in tumor cells21 and because NF-κB is constitutively activated in myeloma cells by environmental stress signals, which is critical to apoptosis evasion and the disease progression of myeloma22,23, we further hypothesized that the SD MVs enriched in the ZNF224 protein enhanced the activity of NF-κB in neighboring RPMI 8226 cells. First, we confirmed the increased activity of NF-κB in RPMI 8226 cells under SD (Figure 7C and 7D), which was consistent with a report that cell stress stimulates NF-kB activation and decreases pro-death signal-induced apoptosis24. Thus, excreting ZNF224 by shedding MVs did not decrease the activity of the NF-κB signaling pathway in the parent RPMI 8226 cells. We then performed a co-culture experiment with SD MVs and RPMI 8226 cells to further study the potential influence of ZNF224 selectively excreted via SD MVs on the activity of NF-kB in neighboring tumor cells. Our data suggested that SD MVs significantly increased NF-κB activation in target RPMI 8226 cells, whereas non-SD MVs did not (Figure 7A and 7B). In addition, we used heat inactivation to inactivate the proteins in SD MVs (HI-SD MVs) and found that these HI-SD MVs failed to activate NF-κB in target tumor cells (Figure 7A and 7B), suggesting a vital role for heat-sensitive components, such as proteins, and especially ZNF224, in the regulation of NF-κB activity in target RPMI 8226 cells.
Activity of NF-κB signaling pathway in the parental and neighbouring RPMI 8226 cells. (A–B) Co-culture of MVs (100 μg/mL) generated by RPMI 8226 cells under SD significantly increased NF-κB activation of the target RPMI 8226 cells while non-SD MVs did not, and heat-inactivation (100 °C for 10 min) of MVs eliminated this effect. This was studied by EMSA. (C–D) The densitometric analysis data of the corresponding EMSA assay for the parental RPMI 8226 cells under SD and non-SD, respectively, suggests increased activity of NF-κB under SD. (E–F) VEGF and IL-6 were selected to be investigated as downstream target genes of NF-κB, and quantitative real-time PCR were performed to confirm the mRNA expression of them at 12 h and 24 h. Data represents the mean±SEM from three independent experiments performed in triplicate. aP>0.05, cP<0.01.
In addition, we examined how the levels of VEGF and IL-6, the two genes that play key roles in myeloma and are regulated by NF-κB, changed in RPMI 8226 cells confronted with SD MVs. SD MVs significantly up-regulated the gene expression of VEGF and IL-6 in target myeloma cells treated with MVs after 12 h and 24 h, respectively; however, this effect was abolished by heat inactivation of SD MVs (Figure 7E and 7F). The results of semiquantitative RT-PCR are shown in Supplementary Figure S1. These data further supported the regulation of NF-κB activity in target RPMI 8226 cells by SD MVs.
SD MVs enhance tube formation of Eahy926 endothelial cells
Angiogenesis is another hallmark of myeloma3. Previously, we found that RPMI 8226 cell-derived MVs significantly enhanced angiogenesis (data not shown). To further investigate the effect of SD MVs on angiogenesis, we tested and compared the effects of non-SD MVs and SD MVs on angiogenesis. Both MVs had an effect on angiogenesis; however, non-SD MVs manifested a notably enhanced capacity to promote in vitro tube formation by Eahy926 endothelial cells (Figure 8A and 8B). To further investigate whether this effect was due to the proteins expressed in SD MVs, we used HI-SD MVs as a control (Figure 8C and 8D). The data showed that HI-SD MVs failed to exert this effect (Figure 8E), which suggested that the proteins selectively expressed in SD MVs were involved in the enhanced angiogenesis of endothelial cells induced by SD MVs.
MVs generated by RPMI 8226 cells under SD promoted tube formation of Eahy926 cells and heat-inactivation of MVs eliminated this effect. (A and B) Typical morphology of the tube-like structure formed by Eahy926 cells (×200) incubated in Matrigel in the presence of 100 μg/mL non-SD MVs and SD MVs for 12 h, respectively. (C and D) The effect of heat-inactivation (HI) (100 °C for 10 min) of MVs on tube formation of Eahy926 cells. (E) Quantification of the number of closed tubes and the comparative results for (A) vs (B) and (C) vs (D), respectively. The results are shown as mean±SEM from three independent experiments. aP>0.05,bP<0.05, cP<0.01.
Discussion
Previously, it was thought that intercellular communication was mediated only by cell-to-cell contact, the release of soluble molecules and gap junctions. In contrast to these well-described mechanisms, MVs began to manifest as a new form of intracellular communication pathway. These vesicles can interact with target cells either by binding via specific receptors or by direct fusion with the plasma membrane25. Tumor cell-derived MVs have been reported to reflect the malignant potential of tumor cells. Increasing evidence suggests that tumor cell-derived MVs play a pivotal role in the development, growth and propagation of a tumor11. Here, we studied various differences in the MV populations that myeloma cells produced under well-fed (10% FBS) and SD (1% FBS) conditions. Our data suggest that SD MVs reflect the SD status of the parent myeloma cells and manifest as the potential driver of SD-dependent upregulation of the anti-apoptotic pathway and angiogenesis during tumor development.
MV shedding is a physiological process that many, if not all, cell types exhibit, and MVs can be detected in various bodily fluids in all individuals. Tumor cells shed MVs more frequently than do normal cells, and the frequency has been shown to correlate with the invasiveness of tumor cells both in vitro26 and in vivo27. Dainiak et al have reported that SD diminished MV formation in B lymphocytes from healthy volunteers28, but the effect of SD on MV formation in tumor cells is still unknown. In this study, we observed a 2.5-fold increase in the number of MVs generated by RPMI 8226 myeloma cells under SD compared with cells that had not undergone SD. Studies have demonstrated that metabolic energy is required for the exfoliation of cell membranes25. It seems that myeloma cells have an intrinsic ability to support intense MV shedding while reprogramming their metabolism to meet the requirements induced by the stresses imposed by nutrient deprivation. Similarly, Wysoczynski et al reported that hypoxia and irradiation induce an increased release of MVs from lung cancer cells29. Thus, tumor cells can be stimulated to release more MVs in adverse tumor microenvironments, and those MVs are potential effectors that can rebuild the microenvironment and rescue the tumor cells. The process of MV formation remains unclear. Studies in dendritic cells and microglia have indicated that MV formation is Ca2+ dependent30,31, and activation of protein kinase C (PKC) is also involved in other cell types32. Reports have shown that GTPase activation induces intense MV release33. However, whether the elevated level of SD MVs with selectively enriched proteins is induced via these mechanisms has yet to be studied.
Although it was reported that stimulation of cells did not result in changes in the size distribution of MVs9, we observed that SD resulted in a different size distribution of MVs compared with non-SD conditions. We found that MVs with diameters below 0.5 μm significantly increased in number after SD treatment, whereas MVs larger than 0.5 μm decreased in number. MVs are heterogeneous in their size and shape, including MVs that are generated by the same parent cells25; however, why and how myeloma cells generate MVs with different sizes under SD remain to be determined.
The proteins in MVs are not randomly released by the parent cells. The composition of MVs seems to result from a special selection process, and the function of MVs appears to be dependent on the cargo that they carry25. P-glycoprotein, a multidrug-resistant (MDR) drug efflux pump, can be transferred to drug-sensitive cells from drug-resistant cells by tumor cell-derived MVs and can turn the drug-sensitive cells into resistant cells34. Another report showed that the oncogenic receptor EGFRvIII, which is exclusively expressed in a subset of aggressive gliomas, can be transferred to a non-aggressive population of tumor cells and can malignantly transform these cells via MVs35. In the present study, we also focused on the proteins in SD MVs and wanted to determine the specific proteins in myeloma.
We identified 3 proteins, ZNF224, SARM, and COBL, that were selectively up-regulated in SD MVs. Among these proteins, the zinc finger transcription factor (ZNF224) is a known transcriptional repressor that can inhibit the transcription of A20, a potent inhibitor of the NF-κB signaling pathway21. NF-κB is activated in myeloma cells under environmental stress, which is central to disease progression, including apoptosis resistance23. Therefore, ZNF224 was chosen for further analysis in the present study. Our results indicate that intense shedding of SD MVs and the enriched ZNF224 protein may act as key factors that affect the activation of NF-κB in neighboring myeloma cells, which may be involved in the anti-apoptotic effects of RPMI 8226 cells under SD. This adaptive stress response may explain the activation of survival cascades and anti-apoptotic mechanisms in acquired resistance to tumor treatments. SARM and COBL, the other two enriched proteins in SD MVs that were identified in our study, also play key roles in the regulation of target tumor cells. SARM has been demonstrated to be an inhibitor of the mTOR signaling pathway, and mTOR activity has been demonstrated to be reduced following SD in many cell types, which results in a decrease in protein synthesis7. The selective excretion of SARM reflects a reorganization of metabolic pathways and the production of biosynthetic intermediates in myeloma cells under SD. The protein COBL is a member of the filament nucleator family, and the upregulation of COBL suggests activation of cytoskeleton reconstitution or MV formation, implying an underestimated role in MV formation. The underlying roles of these selectively enriched proteins in SD MVs are poorly characterized.
The NF-κB signaling pathway plays a critical role in myeloma oncogenesis and progression, including treatment-related resistance. In the present study, we found that SD-induced stress stimulates RPMI 8226 cells to shed more MVs, which were demonstrated to significantly activate NF-κB signaling in target tumor cells, suggesting potential roles in disease progression under SD or other nutrition-deficient conditions. Moreover, SD MVs enhanced the microtubule formation capacity of Eahy926 endothelial cells, which is another important process involved in MM pathogenesis3. However, heat inactivation elicits the stimulatory effect of SD MVs on NF-κB activation and microtubule formation. Because heat inactivation has been recognized as a simple way of denaturing a protein, resulting in the loss of most of its original tertiary structure and its natural bioactivity13, these data suggest a vital role for protein composition in the biological function of SD MVs.
Our data support myeloma cell adaptation to nutrient deprivation-induced stress by activation of the anti-apoptotic pathway and the angiogenic process through shedding of MVs, which suggests an underestimated mechanism in myeloma progression and treatment resistance. Additionally, the selectively sorted proteins may play a key role in the process. Notably, the cargo of MVs, including miRNA, DNA and other biologically active molecules, has been proven to be horizontally transferred to target cells and to participate in the regulation of target cells. This finding indicates that many other molecules are possibly also involved. Another point to be noted is that NF-κB in both the parent and the target myeloma cells was enhanced under SD, accompanied by enhanced levels of the ZNF224 protein in SD cells and MVs. This phenomenon suggests possible involvement of the ZNF224 protein in the regulation of NF-κB activity in both the parent and the target myeloma cells.
The present study unveiled different characteristics of MVs from RPMI 8226 cells generated in response to SD stimulation to gain insights into the biology of MVs in disease progression and treatment resistance. We have found that SD MVs reflect the SD status of donor myeloma cells, with different levels, size distributions, and selectively enriched proteins that potentially regulate neighboring tumor cells. However, the underlying mechanisms of SD MV formation, their release and how proteins are selectively packed into SD MVs remain to be clarified. We hope that this research will both result in and lead to new knowledge and discoveries of the underlying mechanisms of disease progression or drug resistance and to novel strategies to treat myeloma.
Author contribution
Qiu-bai LI and Ping ZOU were the principal investigators and take primary responsibility for the paper; Li SUN, Hong-xiang WANG, Xiao-jian ZHU, Zhi-chao CHEN, Wei-qun CHEN, and Pin-hui WU performed the laboratory work; and Li SUN, Qiu-bai LI, and Pin-hui WU wrote the paper.
References
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–74.
Qin S, Chock PB . Implication of phosphatidylinositol 3-kinase membrane recruitment in hydrogen peroxide-induced activation of PI3K and Akt. Biochemistry 2003; 42: 2995–3003.
Kyle RA, Rajkumar SV . Multiple myeloma. N Engl J Med 2004; 351: 1860–73.
Minarík J, Scudla V, Ordeltová M, Bacovský J, Zemanová M, Pika T . Assessment of plasmatic cell proliferation and apoptosis indexes in the course of multiple myeloma. Vnitr Lek 2008; 54: 31–5.
Levin VA, Panchabhai SC, Shen L, Kornblau SM, Qiu Y, Baggerly KA . Different changes in protein and phosphoprotein levels result from serum starvation of high-grade glioma and adenocarcinoma cell lines. J Proteome Res 2010; 9: 179–91.
Tavaluc RT, Hart LS, Dicker DT, El-Deiry WS . Effects of low confluency, serum starvation and hypoxia on the side population of cancer cell lines. Cell Cycle 2007; 6: 2554–62.
Pirkmajer S, Chibalin AV . Serum starvation: caveat emptor. Am J Physiol Cell Physiol 2011; 301: C272–9.
Pervaiz S, Cao J, Chao OS, Chin YY, Clément MV . Activation of the RacGTPase inhibits apoptosis in human tumor cells. Oncogene 2001; 20: 6263–8.
Bernimoulin M, Waters EK, Foy M, Steele BM, Sullivan M, Falet H, et al. Differential stimulation of monocytic cells results in distinct populations of microparticles. J Thromb Haemost 2009; 7: 1019–28.
Piccin A, Murphy WG, Smith OP . Circulating microparticles: pathophysiology and clinical implications. Blood Reviews 2007; 21: 157–71.
Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C . Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 2010; 123: 1603–11.
Gelderman MP, Simak J . Flow cytometric analysis of cell membrane microparticles. Methods Mol Biol 2008; 484: 79–93.
Shin M, Han Y, Ahn K . The influence of the time and temperature of heat treatment on the allergenicity of egg white proteins. Allergy Asthma Immunol Res 2013; 5: 96–101.
Codeluppi S, Gregory EN, Kjell J, Wigerblad G, Olson L, Svensson CI . Influence of rat substrain and growth conditions on the characteristics of primary cultures of adult rat spinal cord astrocytes. J Neurosci Methods 2011; 197: 118–27.
Pirkmajer S, Chibalin AV . Serum starvation:caveat emptor. Am J Physiol Cell Physiol 2011; 301: C272–9.
Sekiya M, Adachi M, Takayama S, Reed JC, Imai K . IFN-γ upregulates anti-apoptotic gene expression and inhibits apoptosis in IL-3-dependent hematopoietic cells. Biochem Biophys Res Commun 1997; 239: 401–6.
Dolo V, Li R, Dillinger M, Flati S, Manela J, Taylor BJ, et al. Enrichment and localization of ganglioside G(D3) and caveolin-1 in shed tumor cell membrane vesicles. Biochim Biophys Acta 2000; 1486: 265–74.
Medugno L, Florio F, De Cegli R, Grosso M, Lupo A, Costanzo P, et al. The Krüppel-like zinc-finger protein ZNF224 represses aldolase A gene transcription by interacting with the KAP-1co-repressor protein. Gene 2005; 359: 35–43.
O'Neill LA, Fitzgerald KA, Bowie AG . The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol 2003; 24: 286–90.
Dominguez R . Actin filament nucleation and elongation factors-structure-function relationships. Crit Rev Biochem Mol Biol 2009; 4: 351–66.
Harada Y, Kanehira M, Fujisawa Y, Takata R, Shuin T, Miki T, et al. Cell-permeable peptide DEPDC1-ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells molecular and cellular pathobiology. Cancer Res 2010; 70: 5829–39.
Baud V, Karin M . Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov 2009; 8: 33–40.
Klein B . Positioning NF-κB in multiple myeloma. Blood 2010; 115: 3422–4.
Berger R, Jennewein C, Marschall V, Karl S, Cristofanon S, Wagner L, et al. NF-κB is required for smac mimetic-mediated sensitization of glioblastoma cells for γ-irradiation-induced apoptosis. Mol Cancer Ther 2011; 10: 1867–75.
Cocucci E, Racchetti G, Meldolesi J . Shedding microvesicles: artefacts no more. Trends Cell Biol 2009; 19: 43–51.
Ginestra A, La Placa MD, Saladino F, Cassarà D, Nagase H, Vittorelli ML . The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res 1998; 18: 3433–7.
Ginestra A, Miceli D, Dolo V, Romano FM, Vittorelli ML . Membrane vesicles in ovarian cancer fluids: a new potential marker. Anticancer Res 1999; 19: 3439–45.
Dainiak N, Sorba S . Intracellular regulation of the production and release of human erythroid-directed lymphokines. J Clin Invest 1991; 87: 213–20.
Wysoczynski M, Ratajczak MZ . Lung cancer secreted microvesicles: underappreciated modulators of microenvironment in expanding tumors. Int J Cancer 2009; 125: 1595–603.
Bianco F, Pravettoni E, Colombo A, Schenk U, Möller T, Matteoli M, et al. Astrocyte-derived ATP induces vesicle shedding and IL-1 beta lease from microglia. J Immunol 2004; 174: 7268–77.
Pizzirani C, Ferrari D, Chiozzi P, Adinolfi E, Sandonà D, Savaglio E, et al. Stimulation of P2 receptors causes release of IL-1 beta-loaded microvesicles from human dendritic cells. Blood 2007; 109: 3856–64.
Pilzer D, Fishelson Z . Mortalin/GRP75 promotes release of membrane vesicles from immune attacked cells and protection from complement-mediated lysis. Int Immunol 2005; 17: 1239–48.
Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, et al. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol 2009; 19: 1875–85.
Ambudkar SV, Sauna ZE, Gottesman MM, Szakacs G . A novel way to spread drug resistance in tumor cells: functional intercellular transfer of P-glycoprotein (ABCB1). Trends Pharmacol Sci 2005; 26: 385–7.
Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 2008; 10: 619–24.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (81272624 and 81071943). We thank the Central Lab of Wuhan Central Hospital for the detection and analysis of MVs.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary figure is available at the Acta Pharmacologic Sinica website.
Supplementary information
Supplementary Figure S1
mRNA expression of VEGF and IL-6 confirmed by semiquantitative RT-PCR. (PDF 255 kb)
Rights and permissions
About this article
Cite this article
Sun, L., Wang, Hx., Zhu, Xj. et al. Serum deprivation elevates the levels of microvesicles with different size distributions and selectively enriched proteins in human myeloma cells in vitro. Acta Pharmacol Sin 35, 381–393 (2014). https://doi.org/10.1038/aps.2013.166
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2013.166
Keywords
This article is cited by
-
Synergistic vesicle-vector systems for targeted delivery
Journal of Nanobiotechnology (2024)
-
Enhancing the Cellular Production of Extracellular Vesicles for Developing Therapeutic Applications
Pharmaceutical Research (2023)
-
Different Chronic Stress Paradigms Converge on Endogenous TDP43 Cleavage and Aggregation
Molecular Neurobiology (2023)
-
Comparative Analysis of MSC-Derived Exosomes Depending on Cell Culture Media for Regenerative Bioactivity
Tissue Engineering and Regenerative Medicine (2021)
-
Intricate relationships between naked viruses and extracellular vesicles in the crosstalk between pathogen and host
Seminars in Immunopathology (2018)