Abstract
Blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) is an autosomal dominant condition consisting of congenital dysplasia of the eyelids with a reduced horizontal diameter of the palpebral fissures, droopy eyelids and epicanthus inversus. Two clinical entities have been described: type I and type II. The former is distinguished by female infertility, whereas the latter presents without other symptoms. Both type I and type II were recently mapped on the long arm of chromosome 3 (3q22–q23), suggesting a common gene may be affected. The centromeric and the telomeric limits of this region are well defined between loci D3S1316 and D3S1615, which reside approximately 5 cM apart. Here, we present the construction of a YAC contig spanning the entire BPES locus using 17 polymorphic markers, 2 STS and 28 ESTs. This region of approximately 5 Mb was covered by 31 YACs, and was supported by detailed FISH analysis. In addition, we have precisely mapped the propionyl-CoA carboxylase beta polypeptide (PCCB), the gene mutated in propionic acidemia, within this contig. Apart from providing a framework for the identification of the BPES gene, this contig will also be useful for the future identification of defects and genes mapped to this region, and for developing template resources for genomic sequencing.
Similar content being viewed by others
Introduction
Premature ovarian failure and XX gonadal dysgenesis leading to female infertility have been reported in association with an autosomal dominantly inherited malformation of the eyelids: blepharophimosis-ptosis-epicanthus inversus syndrome (BPES; MIM 110100) [1]. This association distinguishes BPES type I from a second form, BPES type II, in which affected females are fertile and the transmission can be observed through either sex. Recently, a defect responsible for BPES type II has been mapped to chromosome 3q22–q23 [2, 3]. The critical region for the gene location is supported by the observation of a translocation breakpoint in an affected patient, in the interval between loci D3S1615 and D3S1316 [4]. The marker D3S1549 maps between these loci and has also been shown to be linked with the disease in other populations [5]. More recently, we have mapped BPES type I, which includes sterility phenotype in affected females to the same location [6]. To facilitate the cloning of the BPES gene, we decided to construct a YAC contig spanning the region marked by the loci most tightly linked to the disease.
Materials and Methods
A set of 31 YACs, potentially residing in 3q22–q23, was directly derived from the information available in the public databases. The YAC clones were obtained from CEPH (Paris). Their chromosomal location and the presence or absence of chimerism was also established by FISH analysis. Cultures obtained from single colonies were grown in YPD medium. DNA was extracted according to standard procedures and used for FISH and PCR assays. For FISH analysis, human metaphase cells were prepared from phytohemagglutinin-stimulated lymphocytes. Digoxigenin-labeled probes were prepared by random priming, and hybridization was detected with anti-digoxi-genin-fluorescein Fab fragments and anti-mouse Ig-digoxigenin F(ab)′2 fragment. Chromosomes were stained with 4′,6-diamidino-2-phenylindole-dihy drochloride (DAPI).
Seventeen polymorphic markers (D3S1590, D3S3696, D3S1549, D3S1316, D3S1615, D3S3528, D3S3617, D3S1576, D3S3554, D3S3586, D3S1309, D3S3637, D3S3641, D3S3684, D3S2453, G09845, GATA.A4A10) and two STS (D3S2699, G02623) were then analyzed individually and incorporated into the map. In addition, twenty-seven ESTs obtained both from the Whitehead Institute Database/MIT and the Science map [7] (WI7255, WI6268, WI9675, A004D46, WI15160, WI11301, WI9039, SGC31992, WI14619, WI15777, WI13771, WI16892, WI13553, WI12317, IB400 SGC31261, SGC32557, SGC32914, WI14004, WI15706, WI17961, WI6881, WI14084, WI6538, A004I40, A005Y32, stSG10017, A004B03), and DRES41 [8] were assessed for possible inclusion in the map. PCR primers and conditions for all of the markers were as described in the data banks. For DRES41 the following primers were used F-GAATTGTGAGCGGATAAC and R-GTTTTCCCAGTCACGACG in a PCR reaction in which each cycle was characterized by 30 s for each step at 94, 56 and 72°C. To reinforce the link between adjacent loci, inter-Alu probes were generated from individual YACs using a PCR primer with the sequence 5′-GTGAGC-CACCGCGCCCGGCC-3′ as previously described [9]. The products were eluted from agarose gels and subsequently used for hybridization to HindIII digested DNA of all individual YACs. In addition, a cDNA selection clone (GT834) obtained by us from YAC 971C7 (data not shown), was found to be identical to propionyl-CoA carboxylase beta polypeptide gene (PCCB) [10] sequences and precisely mapped within the contig.
Results and Discussion
The results on the exact chromosomal location of each YAC and its integrity are reported in table 1. Two YACs clearly mapped on chromosome 3q22, three either in q22 and q23, while the remaining others were in 3q23. Sixteen out of 28 (58%) of the YACs were chimeric. There was agreement between FISH data and hits reported in the data banks for each YACs, although in most cases our FISH data revealed information on YAC integrity.
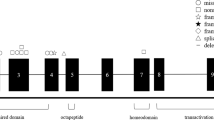
For the construction of the contig, we first focused our attention on D3S1549 and D3S1316 markers. Eighteen YACs were positive for marker D3S1549, while 8 were for marker D3S1316. A compiled summary of the derived YAC contig encompassing the BPES region extended from D3S3586 to D3S1590 is shown in figure 1. It is characterized by 14 polymorphic markers, 1 STS and 1 newly developed inter-Alu PCR probe (MRP1), while eleven ESTs are reported in the upper part of the figure. The order of markers was established by the presence or absence of markers of a single YAC, and by building up the number of overlapping clones. In addition, inter-Alu bands were used as hybridization probes confirming links between adjacent loci. Some YACs contained internal deletions creating ambiguity in the marker order. The order presented assumed the minimum number of internal deletions irrespective of the number of markers missing in each gap. Loci D3S2699, D3S1309, WI13553, WI12317, WI13771, IB400 SGC31261, SGC32557, SGC32914, WI15706, WI17961, WI6881, WI14084, WI6538, A004I40, A005Y32, stSG10017, A004B03 and DRES41 were absent from the presented contig. Marker D3S3637 was present only on one YAC (793F7), while D3S3684 was present on two (793F7 and 942C10), but their exact position remained unsolved.
Physical and genetic map with detailed picture of the YAC contig spanning the BPES critical region. Map is not drawn to scale. YACs are shown below and are represented by a continuous line, except when internal deletions are evident, indicated by dashed lines. Names of clones are those of the corresponding YAC libraries. Loci D3S2699, D3S1309, WI13553, WI12317, WI13771, IB400 SGC31261, SGC32557, SGC32914, WI15706, WI17961, WI6881, WI14084, WI6538, A004I40, A005Y32, stSG10017, A004B03 and DRES41 were absent from the contig, while D3S3637 and D3S3684 being only singly linked to a given YAC were not included in the figure. The arrows indicate the most tightly linked markers and the corresponding BPES type in our sample [3, 6].
The results refined the relative positions of loci reported in the CEPH genetic linkage map [11] and integrated the data of the MIT (contig WC3.18) physical map. Some inconsistencies were detected by comparison with the CEPH linkage map. Markers D3S3641 and D3S3696, positioned 1 cM telomeric to D3S3617 and D3S1615 in the CEPH map, now are inverted and placed centromeric to them. The relative position of the four markers is now solved, being D3S3641, D3S3696, D3S1615, and D3S3617 from the centromere to the telomere. A quite complete concordance with the MIT WC3.18 contig was found. Nevertheless, since our contig is characterized by a high density of YACs between D3S1316 and D3S1615 markers, 15 different additional YACs as compared to WC3.18 contig, the data obtained allow to localize marker D3S1590 centromeric to D3S1615 qnd D3S3696, while D3S3528 occurs telomeric to these loci. The order of the most telomeric markers D3S1316, D3S3554, D3S3586 and GATA.A4A10 is not defined. The exact order of D3S3586 and G09845 could not be resolved, nor that of marker D3S3641 versus D3S1590. The link between D3S1316 and D3S1576 was reinforced by an inter-Alu probe (MRP1) that was present in six YACs (965A1, 943C3, 942H7, 972A7, 942C10, and 946H12). The cDNA selection GT834 clone, identical to PCCB gene, was precisely mapped between markers D3S2453 and D3S3528. This result allowed us to definitely assign propionic acidemia, whose locus was approximately mapped to 3q21–3q22 [10]. A definitive marker order will be better achieved by physical mapping of the YACs resulting in a consensus rare cutter restriction map.
An estimate of the size of the YAC contig was made by summing the sizes of the smallest overlapping and non-overlapping YACs with no known deletions across the contig. By these means, the region covered is approximately 5 Mb. This finding correlates well with the genetic distances of the distant markers of this contig, which should be of about 5 cM [9]. The estimate size of the interval comprised between markers D3S1615 and D3S1316, which narrow the BPES gene region, is of about 2.5 Mb. Seven ESTs are included in this interval, while an additional 4 are immediately outside it. They could be considered as good candidates for BPES.
In conclusion, this contig will be extremely useful for the identification of the BPES gene, which should lead to the understanding of the clinical variation of ovarian failure observed in BPES. This will clarify whether the two forms of the disease are allelic or result as a contiguous gene syndrome. It will also allow the knowledge on how an autosomal gene controls both development and maintenance of ovarian function, features until now thought to be under the control of genes localized on the X chromosome. Moreover, the contig described here will be useful for the future identification of defects and genes mapped to this region. Finally, the expressed genes localized within the contig and the reagents utilized in this work provide the first core of a transcriptional map of this chromosomal region.
References
McKusic VA: Blepharophimosis; in Mendelian Inheritance in Man, ed 10. Baltimore, Johns Hopkins University Press, 1992.
Small KW, Stalvey M, Fisher L, Mullen I, Dickl C, Beadles K, Reimer R, Lessner A, Lewis K, Pericak-Vance M: Blepharophimosis syndrome is linked to chromosome 3q. Hum Mol Genet 1994;54:844–851.
Amati P, Chomel JC, Nivelon-Chevalier A, Gilgenkrantz S, Kitzis A, Kaplan J, Bonneau D: A gene for blepharophimosis, ptosis, epicanthus inversus maps to chromosome 3q23. Hum Genet 1995;96:213–215.
Lawson C, Toomes C, Fryer A, Carette M, Taylor G, Fukushima Y, Dixon M: Definition of the blepharophimosis, ptosis, epicanthus inversus syndrome critical region at chromosome 3q23 based on the analysis of chromosomal anomalies. Hum Mol Genet 1995;4:963–967.
Messiaen L, Leroy B, De Bie S, De Pauw K, Van Roy N, Speleman F, Van Camp G, De Paepe A: Refined genetic and physical mapping of BPES type II. Eur J Hum Genet 1996;4: 34–38.
Amati P, Gasparini P, Zlotogora J, Zelante L, Chomel JC, Kitzis A, Kaplan J, Bonneau D: A gene for premature ovarian failure associated with eyelid malformations maps to chromosomes 3q22-q23. Am J Hum Genet 1996;58: 1089–1092.
Schuler GD, Boguski MS, Hudson TJ, et al: The human gene map. Science 1996;274:540–546.
Banfi S, Borsani G, Rossi E, Bernard L, Guffanti A, Rubboli F, Marchitiello A, Giglio C, Coluccia E, Zollo M, Zuffardi O, Ballabio A: Identification and mapping of human cDNAs homologous to Drosophila mutant genes through EST database searching. Nature Genet 1996;13:167–174.
Totaro A, Grifa A, Roetto A, Lunardi C, D’Agruma L, Sbaiz L, Zelante L, De Sandre G, Camaschella C, Gasparini P: New polymorphisms and markers in the HLA class I region: Relevance to hereditary hemochromatosis. Hum Genet 1995;95:429–434.
Lamhouwah AM, Borankiewicz TJ, Willard HF, Mahuran DS, Gram F, Gravel RA: Isolation of cDNA clones coding for the alpha and beta chains of human propionyl-CoA carboxylase: Chromosomal assignments and DNA polymorphisms associated with PCCA and PCCB genes. Proc Natl Acad Sci USA 1986;83: 4864–4868.
Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal P, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morisette J, Weissenbach J: A comprehensive genetic map of the human genome based on 5,264 micro-satellites. Nature 1996;380:152–154.
Acknowledgments
This work was supported by grant from Telethon (E.469) to L.B., from the Italian Ministry of Health, and from Fédération des Aveugles et Handicapés Visuels de France (P.A.). H.D. and R.M.G. were supported by grant R01HG000358. The authors thank Dr. Banfi (TI-GEM, Milan) for providing DRES41 clone and primers. J.M.R. is a scholar of the Medical Research Council of Canada.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Piemontese, M.R., Memeo, E., Carella, M. et al. A YAC Contig Spanning the Blepharophimosis-Ptosis-Epicanthus inversus Syndrome and Propionic Acidemia Loci. Eur J Hum Genet 5, 171–174 (1997). https://doi.org/10.1007/BF03405896
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03405896