Abstract
The partitioning-defective 3 (Par3), a key component in the conserved Par3/Par6/aPKC complex, plays fundamental roles in cell polarity. Herein we report the identification of Ku70 and Ku80 as novel Par3-interacting proteins through an in vitro binding assay followed by liquid chromatography-tandem mass spectrometry. Ku70/Ku80 proteins are two key regulatory subunits of the DNA-dependent protein kinase (DNA-PK), which plays an essential role in repairing double-strand DNA breaks (DSBs). We determined that the nuclear association of Par3 with Ku70/Ku80 was enhanced by γ-irradiation (IR), a potent DSB inducer. Furthermore, DNA-PKcs, the catalytic subunit of DNA-PK, interacted with the Par3/Ku70/Ku80 complex in response to IR. Par3 over-expression or knockdown was capable of up- or downregulating DNA-PK activity, respectively. Moreover, the Par3 knockdown cells were found to be defective in random plasmid integration, defective in DSB repair following IR, and radiosensitive, phenotypes similar to that of Ku70 knockdown cells. These findings identify Par3 as a novel component of the DNA-PK complex and implicate an unexpected link of cell polarity to DSB repair.
Similar content being viewed by others
Introduction
Cell polarity is vital for the development of multicellular organisms and for the proper functions of epithelial cells in different organs. Polarity in epithelial cells is characterized by different lipid and protein components in the apical and basolateral surface domains. Tight junctions (TJs) act as occluding barriers to maintain cell polarity and homeostasis, and regulate permeability among epithelial cells 1. Three major TJs protein complexes, Crumbs complex, Scribble complex and Par3-Par6-aPKC complex, play important roles in the assembly of TJs 2. Moreover, there is increased evidence indicating that these complexes and their associated proteins may mediate two major types of signals: signals relayed from the intracellular compartments towards TJs regulating their assembly and function; and signals propagated from the TJs into the cells modulating gene expression, cell proliferation and differentiation as well as other functions 2, 3.
The Par3/Par6/aPKC complex is an evolutionarily conserved regulator of cell polarity that plays a central role in forming and maintaining TJs in vertebrate epithelial cells and in specifying neuronal polarity and also in determining asymmetric cell division 4, 5, 6, 7, 8. The Drosophila orthologue of Par3, Bazooka, mediates the asymmetric localization of inscuteable in neuroblast asymmetric cell division and colocalizes with DaPKC and DmPar6 in the apical cortex 9. Thus, it appears that the function of Par3 in cell polarity is conserved across evolution. Additionally, recent studies indicate that Par3 may function in other biological contexts. For instance, over-expression of Par3 in 3T3-L1 adipocytes was found to inhibit insulin-stimulated glucose uptake and insulin-dependent translocation of the glucose transporter GLUT4 to the plasma membrane 10. ASIP-sa (an alternatively spliced form of Par3 with the aPKC binding site) and its interaction with aPKC might contribute to malignant growth and the blocking of Fas/FasL-mediated apoptosis, while ASIP-sb (an alternatively spliced form of Par3 without the aPKC binding site) might function as an antagonist of ASIP-sa 11. In addition, the study by Gao et al. 12 indicates that a fraction of Par3 does not complex with Par6/aPKC and functions as a “free” form in the cytosol. Taken together, these results suggest that Par3 might play additional roles besides establishing cell polarity.
Par3 consists of a conserved region in its N-terminus, three PDZ domains in the middle and the aPKC binding site in the C terminus. PDZ domain is an important module for signal transduction in cells 13. Because Par3 may conduct numerous different cellular tasks, we explored potential Par3-binding partners using a glutathione S-transferase (GST) pull-down assay with the three Par3 PDZ domains, PDZ1, PDZ2 and PDZ3, fused to GST as baits. We report here the identification of 70-kDa (Ku70) and 80-kDa (Ku80) heterodimeric subunits of the DNA-dependent protein kinase (DNA-PK) as Par3-interacting factors. Ku70 and Ku80 interact with the DNA-PK catalytic subunit (DNA-PKcs) to form the DNA-PK complex that regulates the repair of DNA double-strand breaks (DSBs) 14, 15. Subsequent functional analyses revealed a substantial role for Par3 in DSB repair. Our findings suggest that Par3 may function as a new component in the DNA-PK complex and implicate an unexpected link between cell polarity and DSB repair.
Materials and Methods
Cell lines and reagents
A431, Caco2, HeLa and human embryonic kidney 293T cell lines were from ATCC (USA). Minimum essential medium (MEM), Dulbecco's modified Eagle's medium (DMEM) and newborn calf serum were from Gibco (USA). Lipofectamine 2000 was purchased from Invitrogen (Life technologies, USA). Etoposide was purchased from Sigma. Proteinase inhibitor cocktail tablets inhibit a broad spectrum of serine and cysteine proteases were from Roche (USA).
Cell culture
All cell lines used were cultured in MEM or DMEM supplemented with 10% newborn calf serum. For the exogenous expression of proteins in the 293T cells, we used the calcium-phosphate-mediated transfection method. In the HeLa cells, we conducted the transient transfections with Lipofectamine™ 2000 according to the manufacturer's instructions.
Constructs
The full-length Ku70 and Ku80 cDNAs were obtained by amplifying the Ku70 and Ku80 genes from a human liver cDNA library using the following primers: 5′-CGG ATC CAG CCA ACA TGT CAG GGT GGG AG-3′ (Ku70 forward), 5′-CGG GGC CCG TCC TGG AAG TGC TTG GTG AGG G-3′ (Ku70 reverse), 5′-CGG AAT TCA TGG TGC GGT CGG GGA ATA AGG-3′ (Ku80 forward), and 5′-GCT CTA GAT ATC ATG TCC AAT AAA TCG TCC ACA TC-3′ (Ku80 reverse). We cloned the full-length Ku70 and Ku80 into the pcDNA3VSV plasmid (gifted from Axel). The Par3 180K plasmid was kindly provided by Dr Ian G Macara (University of Virginia, USA).
The constructs encoding GST fusion proteins of the ZO-1 PDZ1 domain, N-terminal region (1-273 aa) and C-terminal region (400-609 aa) of Ku70 were generated by subcloning the fragments in-frame into pGEX-5X-1. The C terminus (LCT, position 1 111-1 353 aa) of Par3 long form (180 kDa form) (gifted from Dr Macara, USA) was amplified and then inserted into pGEX-5X-3. PDZ2-3 domain of PTPBAS (1 327-1 612 aa), Par3 PDZ1 domain (251-385 aa), Par3 PDZ2 (425-549 aa), Par3 PDZ2 plus B23 linker (PDZ2+, 425-595 aa), Par3 PDZ3 (577-694 aa), Par3 PDZ3 plus B23 linker (PDZ3+, 531-694 aa), Par3 B23 linker (B23 531-595 aa) and Par3 PDZ2-3 (425-695) were amplified and inserted in-frame into pGEX2T (Amersham Pharmacia Biotech). The His-tagged PDZ2-3 domains of Par3 were generated by inserting the PDZ2-3 domain into pET-3E-His (Novagen, Germany).
GST fusion proteins were produced in Escherichia coli DH5α or BL21DE, and purified on glutathione sepharose 4B using a standard procedure. The His-tagged fusion proteins were expressed from BL21DE and purified on TALON Metal Affinity Resin (Clontech, USA), according to a standard protocol. The sequences of the Par3 siRNA duplexes (siRNA1: GAAACGAAAGCAGAAGAUG, siRNA2: AGGUGAUAAGACUGAUAGA), GFP siRNA control (GFP siRNA: GCAAGCTGACCCTGAAGTTC) and Ku70 (siRNA1: sense GCUCUGCUCAUCAAGUGUCUG; siRNA2: UCCUUGACUUGAUGCACCUGA; siRNA3: ACGGAUCUGACUACUCACUCA; siRNA4: sense ACGAAUUCUAGAGCUUGACCA) were synthesized by Genechem (China). HeLa cells were twice co-transfected with the indicated siRNAs to suppress proteins expression.
Antibodies
The Ku70, Ku80 and DNA-PKcs monoclonal antibodies were purchased from Santa Cruz (Santa Cruz, USA). The Par3 rabbit antisera (anti-Par3LCT and anti-Par3 PDZ2-3 antibodies) were generated against the GST fusion proteins of the Par3 PDZ2-3 and the C terminus, respectively, which were expressed by the aforementioned GST constructs. The Par3 C terminal antisera were first incubated with purified GST proteins bound to glutathione beads, and then were purified by GST-Par3/LCT antigen immobilized on glutathione beads (anti-Par3LCT). The Ku70 rabbit antisera were generated against the GST fusion proteins of the Ku70 N and C terminus expressed by the GST-tagged constructs. The Ku80 rabbit antisera were produced against the GST fusion proteins of the Ku80 C terminus (554-732 aa). The goat anti-mouse and goat anti-rabbit secondary antibodies were purchased from Bio-Rad (USA). The Cy2- and Cy3-conjugated goat anti-mouse antibodies were purchased from Jackson ImmunoResearch Laboratories (USA). The polyclonal and monoclonal DNA-PKcs antibodies against pT2609 were from Dr Benjamin Chen and David Chen (Southwestern Medical Center).
In vitro binding assay
Cells were lysed with 1 ml of lysis buffer (50 mM HEPES pH 7.5, 1% Triton X-100, 150 mM NaCl, 1 mM NaF, 100 μmol PMSF and Cocktail) and incubated with 3 μg purified GST fusion proteins conjugated to glutathione 4B beads. The beads were washed three times with HNTG buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.1% Trition-X-100, 10% glycerol) and eluted with SDS sample buffer. Proteins were separated by SDS-PAGE and analyzed by Coomassie blue staining.
Nuclear fractionation
This method was adapted from a previous protocol 16. Briefly, the HeLa cells were supplemented with hypotonic buffer (10 mM Tirs-Cl, 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, 1 mM NaF (pH 7.2) and Cocktail) and were scraped off and passed through a 25-g needle five times, and centrifugated at 1 000 r.p.m. for 10 min at 4 °C. The resulting crude nuclear pellets were suspended with cell lysis buffer Ι (50 mM HEPES (pH 7.5), 10% glycerol, 0.5% Triton X-100, 150 mM NaCl and Cocktail) and centrifugated at 13 000 r.p.m. for 60 min at 4 °C. The final nuclear pellets were dissolved with RIPA buffer (50 mM HEPES (pH 7.5), 1% Triton X-100, 0.1% SDS, 150 mM NaCl, 1% deoxycholatic sodium, 1 mM NaF and Cocktail) and sonicated on ice.
Immunoprecipitation and immunoblotting
Cells were lysed with 1 ml RIPA buffer. The lysates were quantified using the Bradford assay and were subjected to immunoprecipitation with 3 μl anti-sera. The lysates were incubated with protein A beads overnight at 4 °C, and the beads were washed 4-6 times with HNTG and eluted with SDS loading buffer. The proteins were separated by one-dimensional SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. The antibodies were detected by enhanced chemiluminescence. The densities of western bands were measured with Quantity One software (Bio-Rad, USA).
Immunofluorescence
Cells grown on glass cover slips (Fisher, USA) were washed several times with ice-cold PBS and fixed with −20 °C methanol for 3-5 min. The cells were then blocked in TBST containing 1% BSA for 1 h at room temperature. Antibody incubations were performed at 37 °C for 2 h in TBST containing 1% BSA. The secondary antibodies used were Cy2-conjugated goat anti-rabbit or Cy3-conjugated goat anti-mouse IgG. Cover slips were mounted using PERMAFLUOR aqueous mounting medium (Immunotech, France) and analyzed with a laser-scanning confocal microscope (Leica, Germany).
Ionizing radiation and etoposide treatment
For irradiation, the cells were exposed to indicated Gy of γ-irradiation at a rate of 0.98 Gy/min and recovered in a 37 °C, humidified incubator for the indicated time. A Gammacell-40 exactor (Nordion, Canada) containing a 137Cs was used as an ionizing radiation source. Etoposide was added to the DMEM medium for 1 h, and the cells were lysed and used in the immunoprecipitation assays.
Random plasmid integration
Random plasmid integration was carried out by transfection of the linearized pEGFPC1 expression plasmid. Twenty-four hours later, cells were re-plated at low density in G418-containing medium, and colonies were counted when the control non-transfected cells were all dead.
DNA-PK assay
Whole-cell extracts (WCE) were prepared using the method described by Allalunis-Turner et al. 17. The cells were washed with ice-cold PBS, and the WCE were prepared, adjusting the WCE concentration for equal protein content. DNA-PK Assay Kit was from Promega (USA), and the subsequent steps of the assay were carried out as described by the manufacturer's protocol.
Pulsed-field gel electrophoresis and colony formation assays
Pulsed-field gel electrophoresis (PFGE) was carried out as described before 18. Cells were harvested and about 1×105 cells were embedded in 1% low-melting agarose. Plugs were incubated overnight at 55 °C in 3 ml lysis buffer (50 mM Tris (pH 8.0), 50 mM EDTA (pH 8.0), 2% sarcosyl, 2 mg/ml proteinase K) and washed five times with 3 ml of TE at room temperature. Electrophoresis of the prepared samples was performed using the CHEF-DR® II Pulsed Field Electrophoresis Systems (BioRad, USA). Electrophoretic conditions were as indicated. The colony formation assay was performed as described previously 19.
Results
Ku70/Ku80 specifically interacts with Par3 in vitro and in vivo
To identify proteins that interact with Par3 PDZ domains, GST fusion proteins of PDZ1 and 2-3 domains of Par3 (Supplementary information, Figure S1A) were generated and employed in pull-down assays using lysates of HeLa and A431 cells. The proteins captured by the GST fusion proteins were resolved by SDS polyacrylamide gel electrophoresis (SDS-PAGE). As detected by Coomassie blue staining, two proteins with the molecular weights of 70 and 80 kDa co-purified specifically with the GST-PDZ2-3 fusion protein, but not with the GST control or the GST-PDZ1 protein (Supplementary information, Figure S1B and data not shown). The two bands were excised and analyzed further by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Thirteen peptides were identified, six of which matched Ku70 and seven of which matched Ku80 (Supplementary information, Figure S1B). The mass spectrometry maps of several identified peptides of Ku70 and Ku80 are shown in Supplementary information, Figure S1C.
To confirm the binding of Par3 to the Ku70/Ku80 heterodimer, HeLa cell lysates and A431 cell lysates were mixed with GST or the GST-tagged PDZ2-3 domains of Par3. Monoclonal antibodies against Ku70 and Ku80 detected the presence of these endogenous proteins in the fraction bound to GST-PDZ2-3, but not in that of the GST control (Figure 1A). To verify the interaction between the Ku and Par3 proteins, we transfected constructs encoding vesicular stomatitis virus (VSV)-tagged Ku70 and/or Ku80 into 293T cells. Western blot analysis revealed that the GST- PDZ2-3 fusion protein pulled down Ku70 alone and Ku70/Ku80 together, but not Ku80 alone (Figure 1B), suggesting that Ku70 may mediate the interaction between Par3 and the Ku complex. To test the specificity of the Par3-Ku70 interaction, we constructed several GST-fusion proteins with the PDZ domains derived from the protein tyrosine phosphatase PTPBAS and the tight junction protein Zonula Occludens-1 (ZO-1), which are homologous to the Par3 PDZ domains (Figure 1C). As shown in Figure 1D, only the GST-fusion protein harboring the Par3 PDZ2-3 domains could bind to endogenous Ku70/Ku80 in HeLa cell lysates. These data indicate that the PDZ2-3 domains of Par3 bind specifically to Ku70.
Ku70 and Ku80 bind specifically to the PDZ2-3 domains of Par3. (A) Ku70 and Ku80 associate with PDZ2-3 domains of Par3. A431 cells and HeLa cells were lysed and incubated with GST-tagged PDZ2-3 domains of Par3 bound to glutathione beads. Proteins were detected as indicated. (B) Ku70 mediates the interaction of Ku70/Ku80 with PDZ2-3 domains of Par3. 293T cells transiently transfected with an empty vector, Ku70, Ku80 or Ku70/Ku80 were lysed and incubated with the GST-tagged PDZ2-3 of Par3 bound to glutathione beads, respectively. Proteins were detected as indicated. Aliquots of lysates from the transfected and untransfected cells were run as controls. (C) Alignment of amino-acid sequences of different PDZ domains by the Clustal program. (D) Ku70 and Ku80 specifically associate with PDZ2-3 domains of Par3. HeLa cell lysates were mixed with the indicated GST fusion proteins (1 μg) and the immunoblots were probed as indicated. (E) Association of endogenous Ku70/Ku80 with exogenous Par3. 293T cells were transfected with Par3 as indicated. Proteins were immunoprecipitated from 293T cell lysates and immunoblotted as indicated. (F) The association of Par3 with Ku70/Ku80 occurs endogenously. HeLa cell lysates were immunoprecipitated with antibodies against Ku70 or Par3. Associated proteins were probed as indicated.
To further confirm the physical interaction of Par3 with the Ku complex, immunoprecipitations using antibodies against Ku70 or Ku 80 were performed in the 239T cells transfected with either an empty vector or the construct encoding Par3. As shown in Figure 1E, Par3 co-precipitated with Ku70 and Ku80.
To further document the physiological relevance of the interaction, immunoprecipitation of endogenous Ku70 was conducted in HeLa cells. As shown in Figure 1F, the endogenous Par3 co-immunoprecipitated with the heterodimeric Ku70/Ku80 complex but this did not occur with the control serum. Reciprocally, when endogenous Par3 was precipitated, Ku proteins were also specifically co- precipitated. Since Ku proteins are DNA-binding proteins, to exclude the possibility that DNA mediates the interaction between Par3 and Ku70/Ku80, we treated the Par3 immunoprecipitates with micrococcal nuclease, or added ethidium bromide (EB), which is known to disrupt protein-DNA interactions 20, to the lysates before performing immunoprecipitation. Our data showed that DNA did not mediate Par3-Ku association (Supplementary information, Figure S2). These results clearly demonstrated that Par3 forms a physiological complex with the Ku heterodimer in vivo.
The linker between the PDZ2 and PDZ3 domains within Par3 mediates the interaction with the N-terminus of Ku70
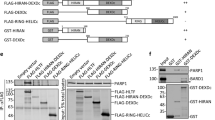
As we demonstrated above, Ku70 interacts with a region in the Par3 PDZ2-3 domains. To pinpoint the region required in PDZ2-3 for this interaction, we generated a Par3 mutant lacking the PDZ2-3 domains (denoted herein as Par3ΔPDZ2-3). Par3 or Par3ΔPDZ2-3 were transfected into 293T cells, and immunoprecipitation was performed using the antibody against Ku70. As expected, Par3 but not Par3ΔPDZ2-3 could interact with Ku70 (Figure 2A), indicating that the Par3 PDZ2-3 region is essential for mediating the Par3 interaction with Ku70. The Par3 PDZ2-3 region is composed of the PDZ2 and PDZ3 domains separated by a linker region, denoted herein as B23. To refine the binding site responsible for the Ku70 interaction within the PDZ2-3 region, various recombinant GST fusion proteins spanning the PDZ2 and PDZ3 domains were generated (Figure 2B). Those GST fusion proteins were incubated with the lysates from HeLa cells for in vitro binding assays. Surprisingly, neither the Par3 PDZ2 nor the PDZ3 domain could bind to Ku70/K80. However, the B23-linker bound specifically to Ku70/Ku80 (Figure 2C). Interestingly, the PDZ2 domain appears to potentiate this interaction, whereas the PDZ3 domain seems to inhibit the binding (Figure 2C, lanes 4 and 6).
Ku70 interacts directly with the Par3 linker region between the PDZ2 and PDZ3 domain via its N terminus. (A) The domain on Par3 responsible for its interaction with Ku70 localizes in its PDZ2-3 domains. 293T cells were transfected with Par3 and Par3ΔPDZ2-3 as indicated. Proteins were immunoprecipitated from 293T cell lysates with antibody against Ku70. Immunoblots were carried out as indicated. (B) Schematic diagram of different regions in Par3 PDZ2 and PDZ3 domains. The binding abilities of the GST fusion proteins to Ku70 are summarized in B (right panel). (C) Par3 interacts with Ku70 via its linker region. The indicated GST fusion proteins were bound to Sepharose beads and then incubated with HeLa cell lysates. The immunoblots were probed as indicated. Total cell lysates were loaded on the right. (D) Schematic representation of Ku70, showing the regions used in GST fusion proteins. (E) His-PDZ2-3 of Par3 binds directly to the N terminal domain of Ku70. Purified PDZ2-3 domains of Par3 carrying the histidine tag was incubated with GST fusion proteins of Ku70 immobilized on Sepharose beads. The associated His-PDZ2-3 was detected with antibody against the histidine tag. Input His-PDZ2-3 was shown on the right. The binding abilities of the GST fusion proteins to Par3 were summarized in (D) (right panel).
Ku70 contains an N-terminal Von Willebrand factor A (VWA) domain, as well as a central Ku70/Ku80 interaction domain and a C-terminal DNA-binding SAP (named after SAF-A/B, Acinus and PIAS) domain. To define the region of Ku70 that mediates the Par3 interaction, GST-Ku70N and -Ku70C fusion proteins were generated (Figure 2D). For the in vitro binding assay, the purified histidine (His)-tagged PDZ2-3 domains of Par3 were incubated with the two GST fusion proteins of Ku70. As shown in Figure 2E, only the Ku70N GST fusion protein containing the VWA domain retrieved the purified His-tagged PDZ2-3 protein, whereas GST alone or the C-terminal Ku70 fusion protein did not. Taken together, our data indicate that the linker B23 region of Par3 mediates a direct and selective binding to the N-terminal region of Ku70, possibly via the VWA domain.
Par3 localizes on the membranes and in the nuclei of HeLa cells
To investigate the biological significance of Par3-Ku70/Ku80 interaction, we first need to know where the interaction occurs in the cells. Ku70/Ku80 mainly localize in the nucleus, playing an important role in DNA repair. To determine the cellular localization of Par3, we conducted sub-cellular fractionation experiments to determine the sub-cellular distributions of Par3 in Caco-2 and HeLa cells. Par3 was clearly detected in both the membrane and nuclear fractions, and at relatively low level in the cytosol by Western blot using the antibody against the Par3 C-terminus (anti-Par3LCT) (Figure 3A). To confirm the sub-cellular distribution pattern of Par3 in Caco-2 and HeLa cells, immunostaining with the purified anti-Par3LCT antibody revealed that the specific signals clearly located to the membrane and in the nucleus (Figure 3B and 3C). Since TJs in most tumor cells are abnormal 21, the membrane signal of Par3 is not in the entire cell-cell junctions as shown by confocal scanning in HeLa cells. These results demonstrate that Par3 is localized in the nuclei, membranes and cytosol in HeLa and Caco-2 cells. The same results were obtained with the purchased Par3 antibodies (Supplementary information, Figure S3A).
Par3 localizes both on the membrane and in the nucleus in HeLa cells. (A) Different sub-cellular fractions of Caco-2 and HeLa cells were prepared. Mem: membrane fraction; Cyto: cytosolic fraction; Nuc: nuclear fraction. Immunoblots were carried out as indicated. (B) The staining pattern using the purified Par3 polyclonal antibodies (anti-Par3LCT) in Caco-2 cells. Caco-2 cells were fixed and stained with the anti-Par3LCT antibodies, and DAPI staining was used to visualize the nuclei of cells. GST-Par3LCT was the antigen used to immunize rabbits to raise the antibodies against Par3. (C) The localization Par3 in HeLa cells. HeLa cells were treated and stained as in (B). The enlarged Par3 signal on the membrane is shown on the left. Scare bar: 10 μm.
γ-irradiation and etoposide enhance interaction of Par3 with Ku proteins
Ku70 and Ku80 were identified as nuclear proteins that play essential roles in DSB repair 22, 23. γ-Irradiation and the radiomimetic chemical etoposide (ETO) are inducers for DSBs. To begin to probe whether the Par3-Ku70 interaction is involved in DSB repair, we monitored their interaction in HeLa and Caco-2 cells irradiated with 10 Gy, and found that the association of Par3 with Ku70 was increased notably in response to γ-irradiation (Figure 4A and 4B). In addition, treating Caco-2 cells with ETO also enhanced the interaction of Par3 with Ku70 (Figure 4C). The data presented herein demonstrate that DNA damage inducers strengthen the Par3-Ku70 interaction, suggesting that Par3 may be involved in DSB repair via its interaction with the Ku proteins upon DSB damage induction. As described earlier, using the EB blocking assay, we excluded the possibility that DNA may play a direct role in mediating the Par3-Ku70 interaction (Supplementary information, Figure S4).
γ-Irradiation positively regulates the binding between Ku and Par3. HeLa cells (A) or Caco2 cells (B) were irradiated with 10 Gy and lysed at the indicated time. Immunoprecipitations and immunoblots were carried out as indicated. (C) Caco-2 cells were treated with 10 μM ETO for 1 h and lysed. Immunoprecipitations and immunoblots were performed as indicated. The data are representative of three repeated experiments. *p<0.05 vs control.
γ-irradiation enhances nuclear localization of Par3, leading to complex formation with the DNA-PK
Since the DNA repair process occurs in the nucleus, we sought to determine whether γ-irradiation affected the nuclear localization of Par3 and subsequently its interaction with the Ku proteins in the nucleus. For these experiments, we performed sub-cellular fractionation assays in HeLa cells treated with or without γ-irradiation. We found that the protein level of Par3 was enhanced in the nuclear fraction in response to γ-irradiation, as compared to that in the control cells (Figure 5A). However, this phenomenon was not observed in the membrane and cytosolic fractions (Figure 5B and 5C). The co-precipitation assays with anti-Par3 demonstrated that γ-irradiation remarkably induced the association of Par3 with Ku70 and Ku80 in the nuclear fraction (Figure 5D). Both Ku and the catalytic subunit of DNA-PK (DNA-PKcs) are involved in DSB repair 24. Interestingly, we detected DNA-PKcs in the immunoprecipitates of Par3 in the nuclear fraction only after IR treatment (Figure 5D), indicating that Par3 complexed with the herterodimer Ku proteins and DNA-PKcs in response to γ-irradiation. To further address this issue, irradiated or non-irradiated HeLa cells were immunostained with Par3 and Ku70 antibodies, and analyzed by confocal microscopy. As shown in Figure 5E, some co-localization of Par3 and Ku70 was observed in the non-irradiated cells (Figure 5E, c). After the ionizing radiation, Par3 in the nucleus was increased notably (Figure 5E, e). The co-localization of Par3 with Ku70 in the nucleus was enhanced when compared to that in the control cells (Figure 5E, g). These results suggest that DSB damage may regulate the nuclear translocation of Par3 and promote its association with the DNA-PK holoenzyme.
γ-Irradiation enhances Par3 nuclear translocation and its co-localization with Ku70. (A–C) HeLa cells treated with or without 10 Gy IR were sub-fractionated. Par3 levels in the three fractions were detected. The data are representative of three independent experiments. *p<0.05 vs control. (D) The nuclear fraction of HeLa cells treated with IR or untreated were prepared and immunoprecipitated with antibody against Par3. Immunoblots were probed as indicated. (E) HeLa cells were untreated or irradiated by 10 Gy and allowed to recover for 2 h after IR. Thereafter, the cells were fixed and stained with Par3 (green) or Ku70 (red) antibodies. Scare bar: 10 μm.
Par3 enhances DNA-PK activity as assayed both in vitro and in vivo
To investigate the biological significance of the Par3-Ku70 interaction, we assessed the effect of Par3 on DNA-PK activity using the in vitro DNA-PK kinase activity assay. HeLa cells were transfected with either an empty vector, wild-type Par3 or Par3ΔB23, which lacks the B23-linker mediating the Par3 interaction with Ku70. WCE were then prepared and assayed for DNA-PK activity in vitro using a commercial kit as described in Materials and Methods. We found that the DNA-PK activity was significantly upregulated in the cells transfected with wild-type Par3 compared to the control cells, while the DNA-PK activity was markedly downregulated by Par3ΔB23 (Figure 6A). The dominant-negative effect of Par3ΔB23 on DNA-PK activity may be due to its ability to form a dimer with the wild-type Par3 25, 26.
Par3 regulates DNA-PK activity as assayed both in vitro and in vivo. (A) DNA-PK activity in cell lysates assayed in vitro is notably upregulated by Par3 over-expression. WCE were prepared from HeLa cells transfected with Par3 or Par3ΔB23, and DNA-PK activities were measured using the DNA-PK Assay System as described in Materials and Methods. The expression levels of Par3 and Par3ΔB23 are shown on the right. *p<0.05. (B, C) Regulation of DNA-PKcs Thr2609 phosphorylation in vivo. HeLa cells transfected with the indicated constructs were mock-treated or irradiated with 10 Gy. Two hours post-irradiation, WCE were prepared and immunoblotted with pT2609. Afterwards, the same blots were stripped and re-probed with the DNA-PKcs monoclonal antibody. The expression of Par3 and Par3ΔB23 were indicated on the right. The data are representative of three repeated experiments. *p<0.05 vs control. (D) Knockdown of Par3 in HeLa cells. HeLa cells were transfected with the two siRNA fragments and lysed 4 days post-transfection. Immunoblots were probed as indicated. (E) Thr2609 phosphorylation of DNA-PKcs is compromised in cells with suppressed expression of Par3. Left: HeLa cells transfected with control siRNA or Par3 siRNA were lysed and immunoprecipitated with monoclonal antibody against DNA-PKcs. Immunoblots were carried out as indicated. Right: Equal amounts of total proteins from cells transfected with control siRNA or Par3 siRNA were loaded and then were immunoblotted with the indicated antibodies.
Currently, the in vivo substrate of DNA-PK remains elusive except for DNA-PKcs itself. Phosphorylation of the DNA-PKcs at Thr2609 is implicated in DNA-PK activation 27, 28, 29. The anti-pThr2609 polyclonal antibodies recognize this modified residue specifically. Thus, anti-pThr2609 antibody is used to monitor DNA-PK activation in vivo. HeLa cells transiently transfected with the pcDNA3 control, Par3 or Par3/ΔB23 constructs were treated with or without 10 Gy γ-irradiation. After a 2-h recovery period, phosphorylation of DNA-PKcs was analyzed by western blot using the anti-pT2609 antibody. IR induced Thr2609 phosphorylation of DNA-PKcs, and the phosphorylation was potentiated by Par3 over-expression while it was clearly attenuated in cells over-expressing Par3ΔB23 (Figure 6B and 6C). These results suggest that the association between Par3 and Ku70 may play a positive role in regulating DNA damage-induced DNA-PK activation.
To further investigate the role of Par3 in DNA-PK activation, we attempted the knockdown of endogenous Par3 by siRNA. The expression of endogenous Par3 in HeLa cells was efficiently suppressed by the Par3 siRNA (Figure 6D). HeLa cells transfected with the Par3 siRNA or control siRNA were treated with or without IR, DNA-PKcs was then immunoprecipitated from these cells and analyzed by western blot using the anti-pT2609 antibody. As shown in Figure 6E, Thr2609 phosphorylation of DNA-PKcs was substantially reduced when Par3 expression was reduced by the specific siRNA as compared to that in the control cells. Taken together, our data indicate that Par3 plays an important in role in DNA-PK activation in response to IR.
Par3 is required for efficient DNA repair and cell survival following DNA damage
DNA-PK-dependent non-homologous end-joining (NHEJ) is a predominant DSB repair pathway in mammalian cells. It is well known that NHEJ is crucial for the efficient random integration of plasmid DNA into the genome of transfected cells 30. As expected, we found that random plasmid integration was markedly impaired in cells downregulated for Ku70 (Figure 7A). Interestingly, knockdown of Par3 also significantly reduced plasmid integration efficiency (Figure 7A), consistent with the notion that Par3 is involved in NHEJ. To further verify this, we performed PFGE to monitor the repair of IR-induced DSBs in vivo. HeLa cells were transfected with siRNAs targeting Par3 or Ku70 or a control siRNA, and after allowing time for protein downregulation, we treated the cells with 70 Gy of IR. HeLa cells transfected with the control siRNA were able to repair most DSBs quickly after irradiation (loss of the faster-migrating DNA signal), while cells downregulated for Par3 or Ku70 displayed higher levels of broken DNA molecules migrating into the gel, and this was the case at both early and later time points following IR (Figure 7B). These data indicate that Par3 is required for efficient DSB repair in HeLa cells.
Par3 knockdown cells are defective in DSB repair and radiosensitive. (A) Downregulation of Par3 or Ku70 causes a marked reduction in random plasmid integration. HeLa cells were transfected with the indicated siRNA oligonucleotides 96 h before transfection with pEGFPC1 (Kan/Neo); 24 h later, the cells were replated at low density in selective medium, and colonies were counted 14 days later. Data represent means ± SD of three independent experiments. (B) Downregulation of Par3 leads to a defect in chromosomal DSB repair. EB-stained PFGE of DNAs in HeLa cells that had been treated with the indicated siRNA oligonucleotides and irradiated (96 h after siRNA transfections). Cells were irradiated with 70 Gy on ice and left for the indicated amount of time at 37 °C to allow for repair. Then 1×105 cells were loaded per well and the gel was run at 120 V, 50–5 000 s linear switch time for 66 h at 14 °C. (C) Temporal analysis of γH2AX foci in Par3 knockdown cells and control cells. Cells transfected with siRNAs targeting Par3 or Ku70 were irradiated with a dose of 1 Gy and stained with γH2AX monoclonal antibody. γH2AX foci of about 100 cells were counted. Bars are mean ± SD of two independent experiments. (D) Survival of Par3 knockdown cells or control cells or Ku70 knockdown cells after IR. Par3 or Ku70 knockdown cells or control cells were irradiated with the indicated dose and were kept in culture for 7 to 10 days before analysis. Bars are means ± SD of three independent experiments.
Phosphorylated H2AX (γH2AX) and quantitative analysis of γH2AX foci following IR are often used for monitoring DSB sites and the efficiency of DNA repair, respectively 29, 31, 32, 33, 34. Our analyses revealed that cells transfected with the control siRNA could repair DSBs efficiently; however, DSB repair in the Par3 or Ku70 knockdown cells was compromised (Figure 7C), further supporting the notion that Par3 positively regulates DSB repair.
To investigate the biological significance of Par3 in DNA damage repair, the radiation sensitivity of Par3 or Ku70 knockdown cells or that of the control cells was examined by assaying their colony-forming ability following IR treatment. As expected, knockdown of Ku70 decreased cell survival substantially after IR, and moreover, the Par3 downregulated cells also displayed significantly reduced survival rates below those of the control cells (Figure 7D). Therefore, Par3 is important for cell survival following IR. Taken together, our functional analyses provide clear evidence that Par3 is required for efficient repair of DSBs and for optimum cell survival following IR-induced DNA damage.
Discussion
In this study, we present a number of independent observations that document a specific interaction between the nuclear protein complex Ku heterodimer and the TJ protein Par3. First, we used GST pull-down assays and LC-MS/MS identification to show that Par3 interacts specifically with Ku70. Second, we determined that Par3 localizes in the nucleus and co-precipitates with Ku from the nuclear extracts of HeLa cells. Moreover, we showed that γ-irradiation and ETO, both of which are potent DNA damage inducers, remarkably enhanced the Par3-Ku70 association. Third, γ-irradiation induces nuclear co-localization of the two proteins in vivo. Finally, the interaction between Par3 and Ku regulates the activity of DNA-PK, which participates in the DSB repair process. These biochemical and morphological observations, as well as our functional analyses, provide compelling evidence of a nuclear complex consisting of Par3, Ku and DNA-PK in different cell types.
Par3 was found to localize in the nucleus under several different experimental conditions in our study (Figure 3B and 3C, and data not shown). Furthermore, immunofluorescence assays detected the nuclear labeling of Par3 in three different cell lines, including Caco-2, HeLa and MDCK, using two different antibodies against two distinct portions of the Par3 polypeptide (Figure 3B and 3C and Supplementary information, Figure S3A). VSV- or GFP-tagged Par3 constructs were clearly detectable in the nuclei of transfected cells (Supplementary information, Figure S3C and S3D). Similar phenomenon of Par3 nuclear localization was also observed by other groups 35, 36. Because this pattern of Par3 was detected with different antibodies and methods, it is highly unlikely that the nuclear staining of Par3 represents non-specific or cross-reactive effects. Moreover, the Par3 nuclear localization appears to correlate with biologically significant events, specifically its interaction with DNA-PK, which was enhanced by γ-irradiation in the nuclear fraction (Figure 5D). γ-Irradiation treatment in HeLa cells significantly increased the co-localization between Par3 and Ku70 in the nucleus (Figure 5E). It seems unlikely that the nuclear localization of Par3 is due to diffusion followed by non-specific trapping within the nucleus, because Par3 is a large protein (180 kDa). Proteins of this size would likely require an active transport process through the nuclear pore complex 37. Sequence analysis of Par3 reveals two K/R-rich stretches of amino acids, KHRK and KKPR (Supplementary Figure S3B), which are similar to the nuclear localization signal (NLS) in simian virus 40 large T antigen, and may serve as putative nuclear localization sequences. There are also two stretches of peptides similar to Xenopus nucleoplasmin NLSs 37. We have not yet determined whether these sequences are responsible for the nuclear enrichment of Par3. However, over-expression of a mutant form of Par3, Par3ΔCT, lacking all the putative NLSs, could still be detected in the nucleus, although at a lower percentage (about 30%) than the wild-type protein (approximate 40%) (Supplementary information, Figure S3C and S3D). It is interesting to note that when expression of a cDNA encoding the last C-terminal 378 amino acids of Par3 (Par3CT), including all four putative NLSs, this mutant form of Par3 is exclusively accumulated within the nucleus (100%; Supplementary information, Figure S3C and S3D). It should also be pointed out that Par3 may not require an NLS of its own, since many nuclear localizing proteins interact with NLS-containing partner proteins to gain nuclear entry via a “piggyback” mechanism 38, 39. Whether Par3 can direct its own nuclear entry or uses an associated partner remains to be determined. Although Ku proteins are predominantly nuclear proteins, they are also reported to localize on the membrane and in the cytosol in different cell lines 40, 41, 42, 43. Although we were able to detect both Ku and DNA-PKcs in the membrane and cytosol fractions using sub-fractionation and western blot assays, Par3 was associated with Ku70/Ku80, but not with DNA-PKcs in these fractions. Thus, the DNA-PKcs does not form a complex with Par3/Ku70/Ku80 on the membrane (data not shown). On the other hand, we observed a clear increase in the association of Par3 and DNA-PK, and detected phosphorylated DNA-PK in the nuclear fraction in response to IR. Along with the data that over-expression or knockdown of Par3 affected DNA-PK activation compared to that in the control cells (Figure 6), we propose that Par3 regulates DNA-PK activity in response to IR in the nucleus.
Par3 functions in DSB repair
In mammalian cells, NHEJ repairs DSBs created by ionizing radiation or during V(D)J recombination. Seven NHEJ factors have been identified: Ku70, Ku80, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Artemis, XRCC4, DNA Ligase IV and Cernunnos/XLF. The Ku70/Ku80 heterodimer forms a ring that preferentially binds to double-strand DNA ends. Ku70/ Ku80 bound to DNA ends recruits DNA-PKcs, which forms a complex with the Artemis nuclease. DNA-PKcs may tether the ends, while Artemis nucleolytically processes DNA ends prior to joining. The Cernunnos/XLF protein forms complexes with XRCC4, DNA Ligase IV, or XRCC4 and Ligase IV simultaneously 44, 45. However, the exact role of Cernunnos-XLF in DNA DSB repair and V(D)J recombination remains to be defined. Our data indicate that DNA damage induced by IR or ETO enhanced the interaction of Ku70 with Par3 (Figure 4), which is consistent with our immunostaining data showing a higher degree of Par3 and Ku co-localization in the nucleus following DNA damage (Figure 5E). In addition, the increased association of Par3 with chromosomal DNA (Supplementary information, Figure S5), and the partial co-localization of Par3 with the pT2609 form of DNA-PKcs and γH2AX in the nucleus in response to IR (data not shown), suggests that Par3 may play a role in DSB repair. This is in line with the observation that over-expression and knockdown of Par3 increase and decrease DNA-PK activity, respectively. Also, Par3 knockdown cells are defective in random plasmid integration, defective in repair of DSBs after IR and radiosensitive. Taken together, these data indicate that Par3, likely through its action on DNA-PK, is involved in DSB rejoining, implicating Par3 as a novel component involved in NHEJ.
Currently the precise role of Par3 in NHEJ pathway is unclear. However, since Par3 is a scaffold protein that plays a key role in establishing cell polarity and TJs, and Ku may serve to tether the free ends of the DSBs to the nuclear matrix 46, it is plausible that Par3 may function as a scaffold to facilitate DNA repair by anchoring the DNA repair apparatus to the broken chromatin. Introduction of DSBs rapidly changes some aspects of higher-order chromatin structure in order to facilitate DNA repair 47, 48, 49. Another potential role of Par3 in DSB repair may be to transmit chromatin-changing signals to DNA-PK and trigger the activation of DNA-PK, which in turn initiates the DNA repair process by phosphorylating its downstream targets.
Intercellular junctions and DNA damage repair
TJ proteins localize on the membrane to take part in TJ establishment and maintenance. There are many lines of evidence indicating that TJ proteins also travel to other sub-cellular locations (including the nucleus) to exert transcriptional/translational or other cellular regulatory activities. For example, the TJ-associated protein Symplekin was reported to participate in nuclear as well as cytoplasmic polyadenylation, suggesting that it may contribute to as yet unexplored functions, such as regulation of mRNA stability and localization 50. TJ protein zonula occluden-2 (ZO-2) regulates TJ assembly and maintenance in quiescent epithelial cells. On the other hand, in sub-confluent MDCK cells, it translocates to the nucleus in proliferating cells, where it may interact with the hnRNP protein SAF-B, as well as bind and inhibit transcription factors AP-1 and C/EBP 51. Interestingly, ZO-1 was also reported to translocate to the nucleus in sub-confluent MDCK and LLC-PK1 cells. However, the biological significance of ZO-1 nuclear translocation remains unclear. In fact, we also clearly detected more Par3 nuclear localization in sub-confluent MDCK cells than the confluent MDCK cells (Supplementary information, Figure S6). In sub-confluent Coco-2 cells, more Par3 in the nucleus was also observed (Figure 3B). Collectively, these data indicated that TJ proteins, such as ZO-2, ZO-1 and Par3, may have distinct functions in different sub-cellular localizations 52. In other words, the data may suggest that epithelial cell polarity may directly connect to nuclear functions through the TJ proteins.
It is worth noting that when cells are in rapid growth or transformation, the polarity of these cells is usually attenuated or even lost 53, 54. Two special isoforms of Par3 may be involved in proliferation and apoptosis in hepatoma cells through unidentified pathway(s) 11. Epithelial-mesenchymal transition (EMT) was defined by the formation of mesenchymal cells from epithelia. Loss or attenuation of epithelial polarity is the hallmark of EMT, which occurs in development and cancer progression 55. The junction proteins localize differently in proliferating (mesenchyma) or TJ-containing (epithelia) cells, which may be reminiscent of their separate functions 56. It is also widely appreciated that the specialized structures at cell contacts (such as TJs, adherent junctions and desmosomes) are focal points for cell-cell signaling pathways implicated in growth and differentiation, and the different roles of the TJ proteins may be achieved through regulating their variant sub-cellular locations.
Sub-confluent cells presumably divide more frequently than confluent cells, and it was also implicated that in the rapidly growing cells DSBs might arise occasionally from closely spaced (single-strand breaks) SSBs or following replication of ROS-induced damage. Also, DSBs might arise when telomeres become critically shortened 44. In addition, NHEJ takes place throughout the cell cycle and is the predominant DSB-rejoining mechanism in the G1 phase 57. Thus, we propose that Par3 may play two distinct roles in regulating the formation of TJs (in TJ-forming cells) on the membrane as well as in participating in repairing the spontaneously occurring DSBs in the nucleus in the different physiological or pathological states of cells.
In fact, intercellular contact has long been considered to be critically important in mediating the cellular response to IR 58, 59. We are interested in learning whether Par3 activities in the nucleus and on the membrane are connected, but we did not observe any detectable Par3 variation in the membrane fraction 2 h following IR (Figure 5B). However, there are indications for TJs' roles in the cellular and pathological processes following exposure to radiation. For instance, TJs in MDCK cells exposed to ionizing irradiation were disassembled 60, 61. Recently, an interesting investigation by Zitzelsberger et al. 62 showed that Par3 gene was strongly amplified in radiation-transformed retinal pigment epithelial cell lines, suggesting that Par3 may function in radiation-induced carcinogenesis. In addition, cellular response to ionizing radiation is cell shape-sensitive 63. It was then noticed that three-dimensional cell contact decreased the radiosensitivity of mammalian cells as compared to those in monolayer cultures. This form of resistance to killing by ionizing radiation was called the “contact” effect 64, 65. Whether the positive regulatory function of Par3 in DSB repair and its role in sustaining cell survival after IR are related to the potential involvement of Par3 in radiation-induced carcinogenesis or contact effect awaits further investigation.
(Supplementary Information is linked to the online version of the paper on the Cell Research website.)
Abbreviations
- (MS):
-
mass spectrometry
- (LC):
-
liquid chromatography
- (MS/MS):
-
tandem mass spectrometry
- (PDZ domain):
-
PSD95/DglA/ZO-1-like domain
- (VWA domain):
-
von Willebrand factor type A domain
- (VSV):
-
vesicular stomatitis virus
- (GST):
-
glutathione S-transferase
- (IR):
-
ionizing radiation
- (DNA-PK):
-
DNA-dependent protein kinase
- (DNA-PKcs):
-
DNA PK catalytic subunit
- (DSBs):
-
DNA double-strand breaks
- (NLS):
-
nuclear localization signal
- (NHEJ):
-
non-homologous end-joining pathway
- (Par3):
-
Partitioning-defective
References
Schneeberger EE, Lynch RD . The tight junction: a multifunctional complex. Am J Physiol Cell Physiol 2004; 286:C1213–C1228.
Matter K, Balda MS . Signalling to and from tight junctions. Nat Rev Mol Cell Biol 2003; 4:225–236.
Balda MS, Garrett MD, Matter K . The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol 2003; 160:423–432.
Izumi Y, Hirose T, Tamai Y, et al. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol 1998; 143:95–106.
Joberty G, Petersen C, Gao L, Macara IG . The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2000; 2:531–539.
Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T . A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol 2000; 2:540–547.
Shi SH, Jan LY, Jan YN . Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 2003; 112:63–75.
Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K . Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol 2004; 6:328–334.
Schober M, Schaefer M, Knoblich JA . Bazooka recruits inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 1999; 402:548–551.
Kotani K, Ogawa W, Hashiramoto M, Onishi T, Ohno S, Kasuga M . Inhibition of insulin-induced glucose uptake by atypical protein kinase C isotype-specific interacting protein in 3T3-L1 adipocytes. J Biol Chem 2000; 275:26390–26395.
Hu Y, Fang C, Xu Y . The effect of isoforms of the cell polarity protein, human ASIP, on the cell cycle and Fas/FasL-mediated apoptosis in human hepatoma cells. Cell Mol Life Sci 2005; 62:1974–1983.
Gao L, Joberty G, Macara IG . Assembly of epithelial tight junctions is negatively regulated by Par6. Curr Biol 2002; 12:221–225.
Sheng M, Sala C . PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 2001; 24:1–29.
Casellas R, Nussenzweig A, Wuerffel R, et al. Ku80 is required for immunoglobulin isotype switching. EMBO J 1998; 17:2404–2411.
Taccioli GE, Gottlieb TM, Blunt T, et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 1994; 265:1442–1445.
Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer SJ . A role for proapoptotic BID in the DNA-damage response. Cell 2005; 122:579–591.
Allalunis-Turner MJ, Lintott LG, Barron GM, Day RS III, Lees-Miller SP . Lack of correlation between DNA-dependent protein kinase activity and tumor cell radiosensitivity. Cancer Res 1995; 55:5200–5202.
Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M . A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res 2004; 64:500–508.
Kurimasa A, Kumano S, Boubnov NV, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol 1999; 19:3877–3884.
Lai JS, Herr W . Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA 1992; 89:6958–6962.
Katoh M . Epithelial-mesenchymal transition in gastric cancer (Review). Int J Oncol 2005; 27:1677–1683.
Jin S, Weaver DT . Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J 1997; 16:6874–6885.
Wu X, Lieber MR . Protein-protein and protein-DNA interaction regions within the DNA end-binding protein Ku70-Ku86. Mol Cell Biol 1996; 16:5186–5193.
Doherty AJ, Jackson SP . DNA repair: how Ku makes ends meet. Curr Biol 2001; 11:R920–R924.
Benton R, St Johnston D . A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol 2003; 13:1330–1334.
Mizuno K, Suzuki A, Hirose T, et al. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem 2003; 278:31240–31250.
Chan DW, Chen BP, Prithivirajsingh S, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 2002; 16:2333–2338.
Wechsler T, Chen BP, Harper R, et al. DNA-PKcs function regulated specifically by protein phosphatase 5. Proc Natl Acad Sci USA 2004; 101:1247–1252.
Falck J, Coates J, Jackson SP . Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005; 434:605–611.
Lou Z, Chen BP, Asaithamby A, Minter-Dykhouse K, Chen DJ, Chen J . MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J Biol Chem 2004; 279:46359–46362.
Rogakou EP, Boon C, Redon C, Bonner WM . Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999; 146:905–916.
Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM . A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 2000; 10:886–895.
Rothkamm K, Lobrich M . Evidence for a lack of DNA double-strand break repair in human cells exposed to very low X-ray doses. Proc Natl Acad Sci USA 2003; 100:5057–5062.
Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA . ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 2004; 64:2390–2396.
Lechler T, Fuchs E . Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 2005; 437:275–280.
Brajenovic M, Joberty G, Kuster B, Bouwmeester T, Drewes G . Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J Biol Chem 2004; 279:12804–12811.
Forbes DJ . Structure and function of the nuclear pore complex. Annu Rev Cell Biol 1992; 8:495–527.
Dingwall C, Sharnick SV, Laskey RA . A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 1982; 30:449–458.
Zhao LJ, Padmanabhan R . Nuclear transport of adenovirus DNA polymerase is facilitated by interaction with preterminal protein. Cell 1988; 55:1005–1015.
Dalziel RG, Mendelson SC, Quinn JP . The nuclear autoimmune antigen Ku is also present on the cell surface. Autoimmunity 1992; 13:265–267.
Monferran S, Muller C, Mourey L, Frit P, Salles B . The membrane-associated form of the DNA repair protein Ku is involved in cell adhesion to fibronectin. J Mol Biol 2004; 337:503–511.
Prabhakar BS, Allaway GP, Srinivasappa J, Notkins AL . Cell surface expression of the 70-kD component of Ku, a DNA-binding nuclear autoantigen. J Clin Invest 1990; 86:1301–1305.
Ginis I, Mentzer SJ, Li X, Faller DV . Characterization of a hypoxia-responsive adhesion molecule for leukocytes on human endothelial cells. J Immunol 1995; 155:802–810.
O'Driscoll M, Jeggo PA . The role of double-strand break repair – insights from human genetics. Nat Rev Genet 2006; 7:45–54.
Gu Y, Seidl KJ, Rathbun GA, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 1997; 7:653–665.
Rodgers W, Jordan SJ, Capra JD . Transient association of Ku with nuclear substrates characterized using fluorescence photobleaching. J Immunol 2002; 168:2348–2355.
Bakkenist CJ, Kastan MB . DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421:499–506.
Shiloh Y . ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev 2001; 11:71–77.
van Attikum H, Gasser SM . The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol 2005; 6:757–765.
Barnard DC, Ryan K, Manley JL, Richter JD . Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 2004; 119:641–651.
Traweger A, Fuchs R, Krizbai IA, Weiger TM, Bauer HC, Bauer H . The tight junction protein ZO-2 localizes to the nucleus and interacts with the heterogeneous nuclear ribonucleoprotein scaffold attachment factor-B. J Biol Chem 2003; 278:2692–2700.
Chlenski A, Ketels KV, Korovaitseva GI, Talamonti MS, Oyasu R, Scarpelli DG . Organization and expression of the human zo-2 gene (tjp-2) in normal and neoplastic tissues. Biochim Biophys Acta 2000; 1493:319–324.
Soler AP, Laughlin KV, Mullin JM . Effects of epidermal growth factor versus phorbol ester on kidney epithelial (LLC-PK1) tight junction permeability and cell division. Exp Cell Res 1993; 207:398–406.
Schoenenberger CA, Zuk A, Kendall D, Matlin KS . Multilayering and loss of apical polarity in MDCK cells transformed with viral K-ras. J Cell Biol 1991; 112:873–889.
Thiery JP . Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol 2003; 15:740–746.
Eger A, Stockinger A, Park J, et al. Beta-Catenin and TGFbeta signalling cooperate to maintain a mesenchymal phenotype after FosER-induced epithelial to mesenchymal transition. Oncogene 2004; 23:2672–2680.
Lieber MR, Ma Y, Pannicke U, Schwarz K . Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol 2003; 4:712–720.
Durand RE, Sutherland RM . Growth and radiation survival characteristics of V79-171b Chinese hamster cells: a possible influence of intercellular contact. Radiat Res 1973; 56:513–527.
Trosko JE, Inoue T . Oxidative stress, signal transduction, and intercellular communication in radiation carcinogenesis. Stem Cells 1997; 15 Suppl 2:59–67.
Somosy Z, Bognar G, Thuroczy G, Koteles GJ . Biological responses of tight junction to ionizing radiation and electromagnetic field expostion. Cell Mol Biol (Noisy-le-grand) 2002; 48:571–575.
Porvaznik M . Tight junction disruption and recovery after sublethal gamma irradiation. Radiat Res 1979; 78:233–250.
Zitzelsberger H, Hieber L, Richter H, et al. Gene amplification of atypical PKC-binding PARD3 in radiation-transformed neoplastic retinal pigment epithelial cell lines. Genes Chromosomes Cancer 2004; 40:55–59.
Reddy NM, Lange CS . Serum, trypsin, and cell shape but not cell-to-cell contact influence the X-ray sensitivity of Chinese hamster V79 cells in monolayers and in spheroids. Radiat Res 1991; 127:30–35.
Sutherland RM . Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 1988; 240:177–184.
Olive PL, Durand RE . Drug and radiation resistance in spheroids: cell contact and kinetics. Cancer Metastasis Rev 1994; 13:121–138.
Acknowledgements
We thank Dr Yingjie Wu for critical reading of the manuscript. We also thank Drs Stephen Jackson, Peter Ahnesorg, Terry Lechler and Gerard Drewes for helpful communication. We thank Dr Ian G Macara for providing the Par3 plasmid. We thank Dr Hu Zhou for the experiment. This work was supported by the grants from National Natural Science Foundation of China (Nos. 30170208, 30623003 and 30170208) and from the Ministry of Science and Technology, China (No. 2001AA233031 and No. 2001CB510205), and from US NIH (CA50519) (to DJ Chen).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Fang, L., Wang, Y., Du, D. et al. Cell polarity protein Par3 complexes with DNA-PK via Ku70 and regulates DNA double-strand break repair. Cell Res 17, 100–116 (2007). https://doi.org/10.1038/sj.cr.7310145
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7310145
Keywords
This article is cited by
-
The polarity protein PARD3 and cancer
Oncogene (2021)
-
Polarity signaling ensures epidermal homeostasis by coupling cellular mechanics and genomic integrity
Nature Communications (2019)
-
Elevated leptin disrupts epithelial polarity and promotes premalignant alterations in the mammary gland
Oncogene (2019)
-
Shared and independent functions of aPKCλ and Par3 in skin tumorigenesis
Oncogene (2018)
-
Dual function of partitioning-defective 3 in the regulation of YAP phosphorylation and activation
Cell Discovery (2016)