Abstract
By reducing cysteine-sulfinic acid in oxidized peroxiredoxin, sulfiredoxin (Srx) plays an important role in oxidation stress resistance in yeast and human cells. Here, we report the first molecular and functional characterization of Srx homolog from higher plants. Bioinformatic analysis revealed the presence of potential Srx encoding sequences in both monocot and dicot plant species. Putative plant Srx proteins exhibited significant identities to their orthologs from yeast and human, and contained the conserved signature sequence and residues essential for catalysis. However, unlike yeast and human orthologs, plant Srxs were all predicted to possess chloroplast transit peptide in their primary structure. The Srx proteins from Arabidopsis and rice (designated as AtSrx and OsSrx, respectively) complemented functional deficiency of Srx in the SRX1 deletion yeast cells. A GFP fusion protein of AtSrx was targeted to chloroplast in Arabidopsis mesophyll protoplast. AtSrx transcription occurred in both vegetative and reproductive organs, and the highest transcript level was detected in leaves. Under oxidation stress, AtSrx transcript level was substantially increased, which paralleled with enhanced transcription of 2-Cys peroxiredoxins that have been found essential in maintaining chloroplast redox balance. In addition to oxidation stress, osmotic/water deficit or cold treatments also raised AtSrx transcript level. Consistent with above findings, the knock-out mutant of AtSrx was significantly more susceptible to oxidation stress than wild type Arabidopsis plant. Taken together, the results of this work indicate the existence of functional Srx homolog in higher plants that is essential for plants to cope with oxidation stress.
Similar content being viewed by others
Introduction
The sulfur atom in cysteine (Cys) is sensitive to oxidation by a variety of oxidants including reactive oxygen species (ROS) that come from environment or are produced as byproduct of cellular metabolism 1, 2. Consequently, the sulfhydryl group (-SH) of cysteine residue in cellular proteins can exist in several different oxidation states 3. Reversible oxidation of cysteine residues to form disulfide bridge (-S-S-), which is an important posttranslational modification and contributes to regulation of protein function, has been studied extensively 4. In contrast, the oxidation of Cys-SH to Cys-SOH (sulfenic acid), Cys-SO2H (sulfinic acid) or Cys-SO3H (sulfonic acid) has not been well investigated 3. While it is known that cysteine disulfide and Cys-SOH are reduced by glutathione and thioredoxin 4, the proteins involved in reducing Cys-SO2H have only begun to be analyzed very recently. In 2003, the yeast protein sulfiredoxin (Srx) was identified with the capacity to reduce Cys-SO2H in overoxidized peroxiredoxin (Prx) Tsa1 in the presence of ATP and Mg2+ 5. Subsequently, the mammalian orthologs of Srx were also found to participate in the reduction of Cys-SO2H, which again involved ATP hydrolysis 6. Surprisingly, an extensive biochemical investigation has shown that the reduction of Cys-SO2H by purified human Srx (hSrx) is specific to 2-Cys Prx isoforms 7, which contain two conserved cysteine residues separated by 121 amino acid residues 8. The high specificity of hSrx catalysis is tightly correlated with the specific binding of hSrx to the 2-Cys Prxs 7.
Amino acid sequence comparison has indicated that Srx proteins from eukaryotic organisms share a similar primary structure and contain the conserved signature sequence Phe-Gly/Ser-Gly-Cys-His-Arg 9. Crystal structure analysis has shown that the overall topology of hSrx is composed of three α helices and one five-stranded, mixed β sheet 9. While helices α1 and α3 are located at the two ends of the structure, helix α2 is positioned internally and close to the β sheet. The signature sequence, Phe96-Gly97-Gly98-Cys99-His100-Arg101, is contained in the N-terminus of helix α2 and the coil structure immediately before this helix. Within this element, the Gly98-Cys99-His100-Arg101 element is directly involved in ATP binding, and Cys99 is the essential active site residue. In addition to the residues located in the signature sequence, Arg51 and Lys61 also contributes to ATP binding, and the two residues have been found conserved in yeast and mammalian Srx proteins described thus far 5, 6, 9. During catalysis, the 2-Cys Prx containing Cys-SO2H binds specifically to hSrx, the sulfinic group is then activated by reacting with ATP, leading to the formation of a sulfinic phosphoryl ester 9. The conversion of sulfinic phosphoryl ester to the reduced sulfhydryl group involves additional enzymes and is thiol-dependent 9.
Compared to yeast and mammals, systematic efforts in studying the molecular and biochemical properties of Srx proteins from higher plants have not been reported. In plant cells, ROS are produced during normal metabolic and developmental processes or in response to biotic and abiotic stresses 10. Chloroplast, which is the site of photosynthesis and home of several important biosynthetic pathways involved in the synthesis of amino acids, fatty acids or nucleotides, is one of the main venues for ROS production in green plant tissues 10. 2-Cys Prx isoforms have been found to play an essential role in the detoxification of peroxides within chloroplast 11. Considering the functional relationship between 2-Cys Prx and Srx that has been demonstrated in yeast and animal cells 5, 6, 12, it is important to examine if there is functional Srx homolog in higher plants and whether plant Srx is targeted to chloroplast. In this work, we provide molecular genetic evidence for the presence of functional Srx homolog in both dicot and monocot plants, and discuss the potential role of plant Srx protein under normal or stress conditions.
Materials and Methods
Plant and yeast materials, PCR primers and general molecular methods
Arabidopsis thaliana (ecotype Col-0) and rice (Oryza sativa L. cv. 'Nipponbare') plants were maintained in greenhouse. The temperatures for Arabidopsis and rice growth were 22 °C and 28 °C, respectively. The standard photoperiod and medium for growing Arabidopsis aseptically are 16 h light/ 8 h dark and MS plate supplemented with 2% sucrose, respectively. Saccharomyces cerevisiae SRX1 deletion strain (MAT alpha; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0;YKL086w::kanMX4) was obtained from EUROSCARF (http://www.uni-frankfurt.de/fb15/mikro/euroscarf/). Chemicals for yeast complementation experiment (hydrogen peroxide) and osmotic/water deficit stress treatment (polyethylene glycol 8000) were purchased from Sigma. The media for growing Arabidopsis seedlings and plants were prepared as previously described 13. Yeast growth media were prepared according to methods detailed in the website of Invitrogen (http://www.invitrogen.com/content/sfs/manuals/pyes2_man.pdf). The oligonucleotide primers used in this work are listed in Table 1. General molecular methods for manipulating DNA or RNA are those detailed previously 14. The correctness of DNA cloning experiments was always confirmed by restriction enzyme digestion analysis and DNA sequencing.
Bioinformatic analysis
Previous study has identified a potential homolog of yeast Srx in Arabidopsis 5. This putative protein (GenBank accession No. AAO42977) is encoded by the locus At1g31170. Using the deduced coding and protein sequences of Arabidopsis Srx homolog as queries, a variety of public nucleic acid and protein databases was searched, which resulted in the finding of potential homologs of yeast Srx in both higher plants and algae species. The identities of putative plant Srxs to their yeast and human orthologs were computed using the MegAlign program of DNAStar (Laser gene). For analyzing conserved elements in primary structure, the deduced amino acid sequences of putative plant Srxs were aligned with those of human and yeast Srx proteins using ClustalW (www.ebi.ac.uk/clustalw/index.html). For predicting chloroplast transit peptide in putative plant Srxs, the ChloroP1.1 and TargetP1.1 programs (http://www.cbs.dtu.dk/services/) were used with default options.
Cloning of putative Srx cDNAs from Arabidopsis and rice
Gene specific primers were designed for amplifying cDNAs corresponding to the coding regions of Srxs from Arabidopsis and rice. Total RNA samples were isolated from Arabidopsis and rice leaves using RNeasy plant mini kit (Qiagen). After further treatment with RNase-free-DNase set (Qiagen) (to remove genomic DNA contamination), the RNA samples were converted into cDNA using M-MLV reverse transcriptase (Promega). PCR experiments using cDNA samples were conducted as detailed previously 15. The cDNA fragment of expected size was cloned using the pGEM®-T Easy plasmid vector (Promega). Positive clones (identified by restriction enzyme digestion analysis and DNA sequencing) were sequenced from both strands. The cDNA sequence of Arabidopsis Srx obtained in this work was identical to a previously submitted mRNA sequence (GenBank accession No. BT004731) that is specified by Arabidopsis locus At1g31170. The cDNA sequence of rice Srx determined in this work was deposited in GenBank with the accession number DQ132851.
Complementation of Srx deficiency in SRX1 deletion yeast cells
To test if putative plant Srx would functionally complement Srx deficiency in yeast cells, the cDNAs of Arabidopsis and rice Srxs were ectopically expressed in the SRX1 deletion yeast strain. The two plant cDNAs were individually cloned into the galactose inducible yeast expression vector pYES2 (Invitrogen), resulting in the constructs pAtSrx and pOsSrx, respectively. The two expression constructs and the control vector pYES2 were each transformed into the SRX1 deletion cells using a simplified yeast transformation method 16. The resulted recombinant strains were selected using uracil deficient medium and were further verified by PCR amplification of AtSrx or OsSrx coding regions 17. The positive colonies and the colony containing pYES2 were streaked onto uracil-free plates that contained 2 % galactose and 2 mM hydrogen peroxide. The plates were then incubated at 28 °C for 72 h before yeast growth was examined and photographed.
Intracellular targeting of putative Arabidopsis Srx
Three DNA constructs (p35S-GFP, p35S-AtSrx-GFP, p35S-mAtSrx-GFP) were prepared for investigating intracellular targeting of Arabidopsis Srx using transient expression in Arabidopsis mesophyll protoplasts. p35S-GFP was created by cloning the coding region of EGFP (Clontech) down-stream of the 35S promoter in the plasmid vector pJIT163 (http://www.pgreen.ac.uk/). p35S-AtSrx-GFP was constructed by cloning the complete coding region of Arabidopsis Srx up-stream (and in frame with) the GFP coding region of p35S-GFP. p35S-mAtSrx-GFP was developed similarly as p35S-AtSrx-GFP, except that the coding region of Arabidopsis Srx in this construct was truncated by lacking the coding sequence for the first 30 amino acid residues. These residues had been predicted to form a potential chloroplast transit peptide by ChloroP1.1 program. Arabidopsis mesophyll protoplasts were isolated 18, transfected with each of above three constructs 19, and examined by dual channel confocal microscopy 20. The bright field image, GFP fluorescence, and the red autofluorescence of chloroplast from expressing protoplast were recorded simultaneously and compared. Potential colocalization of GFP fluorescence with chloroplast autofluorescence was further analyzed by checking the presence of yellow signal in superimposed images.
Examination of AtSrx transcript levels in Arabidopsis organs and during stress
To compare the relative transcript levels of AtSrx in Arabidopsis vegetative and reproductive organs, total RNA samples were isolated from roots, stems, leaves, flowers and siliques, respectively, and were reversely transcribed into cDNAs as described above. The cDNA contents of different reverse transcription mixtures were carefully normalized by amplifying tubulin transcripts 21. The relative transcript levels of AtSrx in the different organs were then evaluated using the normalized cDNA samples. To monitor the kinetics of above semiquantitative RT-PCR, tubulin transcripts were amplified (using the same set of normalized cDNA samples) with different numbers of amplification cycles (i. e., 24, 28, 32, or 35).
For stress treatment, Arabidopsis seedlings (2 weeks old) were transferred into liquid MS medium (supplemented with 1% sucrose). After two day adaptation, oxidation stress was initiated by adding hydrogen peroxide (4 mM final concentration) to the liquid medium 22, 23. The duration of the treatment was 6 h. Osmotic/water deficit stress was augmented by growing the adapted seedlings in liquid MS medium containing 10% PEG8000 for 8 h 24. Cold stress was accomplished by growing the adapted seedlings (in liquid MS medium) at 4 °C for 8 h 25. Following each treatment, the seedlings were immediately harvested and stored in liquid nitrogen. AtSrx transcript levels in the stressed seedlings were evaluated (using semiquantitative RT-PCR) as described above. As positive control for oxidant induced gene expression changes, alterations in the transcript levels of 2-Cys Prx proteins (A and B) in response to the oxidation stress imposed in this work were also investigated.
Characterization of AtSrx knock-out mutant
A T-DNA insertion line, SALK_015324, was identified for AtSrx and obtained from Arabidopsis Biological Resource Center (ABRC, http://www.biosci.ohio-state.edu/). Plants homozygous for T-DNA insertion were identified from T3 population and confirmed in the T4 generation using genomic PCR according to published methods (http://signal.salk.edu/). The lack of AtSrx transcripts in the homozygous T-DNA insertion line (designated as atsrx1-1) was confirmed using RT-PCR with AtSrx specific primers (Table 1).
For comparing the oxidation stress response of atsrx1-1 and wild type plants, seedlings (grown under standard condition, with 6 to 9 rosette leaves) were transferred onto MS medium supplemented with 10 μM paraquat (methyl viologen, Sigma). After growing for a further seven days (under 16 h light/8 h dark photoperiod), the phenotypes of the treated plants were recorded using a digital camera. atsrx1-1 and wild type plants were also tested for difference in maximal quantum yield of photosystem II (Fv/Fm ratio), which is inversely related to the severity of stress 26, 27, before and after oxidation treatment using dark-adapted leaves as described previously 28. For investigating maximal quantum yield before oxidation stress, the mature rosettes of atsrx1-1 and wild type plants (4-week old, grown under standard condition) were transferred to dark. After 20 min in the dark, the fully expanded leaves were used for determining Fv/Fm ratio. For investigating maximal quantum yield after oxidation stress, the mature rosettes of atsrx1-1 and wild type plants (as described above) were sprayed with a solution containing 100 mM paraquat and 0.5% Tween 20 (Sigma). The sprayed plants were kept in dark for 3 h (at 22 °C) before being transferred to standard growth condition (with 16 h light/ 8 h dark photoperiod). Fv/Fm ratio was determined (towards the end of the 8 h dark photoperiod) at 24 h and 48 h after shifting to the normal growth condition, respectively.
As an essential component to above genetic analysis, complementation experiment was conducted to examine if cloned AtSrx cDNA could correct the phenotype of AtSrx knock-out line under oxidation stress. For this purpose, a 35S-AtSrx expression cassette was created by cloning the coding sequence of AtSrx into pJIT163 (see above). This cassette was then ligated into the SacI and SmaI sites of the plant transformation vector pBINPLUS 29, creating the T-DNA construct pBin35S-AtSrx. The T-DNA cassette was introduced into atsrx1-1 background using Agrobacterium mediated transformation 30. Kanamycin resistant T1 plants (with 6 to 9 rosette leaves) and wild type plants of similar developmental stage were transferred onto MS medium containing 10 μM paraquat. The phenotypes of the treated plants were examined and recorded as described above.
Results
Primary structure of putative Srx proteins from higher plants
Prior to this work, the presence of a putative Srx homolog in Arabidopsis (AtSrx) has been noted 5. However, there was no knowledge on Srx in other plant species. By searching public databases using AtSrx as query, potential Srx encoding sequences were found in a variety of plant and algae species. For investigating general features in the primary structure of plant Srx proteins in this work, the deduced amino acid sequences of putative Srxs from five monocot (barley, Sorghum, rice, rye, maize) and three dicot (Arabidopsis, potato, tomato) species were analyzed in more detail. At the amino sequence level, plant Srxs were 21.6-25.0% identical to their yeast ortholog and 19.0-30.3% identical to hSrx. Among the eight plant Srxs, the highest identity score was 99.2% (between potato and tomato Srxs), whereas the lowest one was 58.9% (between tomato and rye Srxs). The signature sequence Phe-Gly/Ser-Gly-Cys-His-Arg was present in all eight plant Srxs, and the Gly-Cys-His-Arg element (within the signature sequence) was strictly conserved (Figure 1, boxed region). Apart from signature sequence, the two residues corresponding to Arg51 and Lys61 of hSrx were also strictly conserved in the eight plant Srxs (Figure 1, indicated by arrowhead). Using ChloroP program, the eight plant Srxs were all predicted to contain chloroplast transit peptide in the N-terminus of their primary structure (data not shown). A similar finding was also obtained when the TargetP program was employed for prediction.
Comparison of the deduced amino acid sequences of Srx proteins from eight plant species, human and yeast (S. cerevisiae). The conserved signature sequence (FG/SGCHR) is boxed. The residues corresponding to Arg51 and Lys61 of hSrx (indicated by arrowheads) were also conserved in plant and yeast Srx proteins. The GenBank accession numbers for Arabidopsis, rice, human and yeast Srx sequences are BT004731, DQ132851, AAH47707 and P36077, respectively. The transcripts, from which the deduced amino acid sequences of the putative Srxs from barley, maize, potato, rye, sorghum and tomato are derived, were identified in TIGR Gene Indices website (http://www.tigr.org/tdb/tgi/plant.shtml). Their accession numbers are TC141756, TC250781, TC97423, TC2742, TC105843 and TC160927, respectively.
Complementation of yeast Srx deficiency by AtSrx or OsSrx
In yeast, functional deficiency of Srx (by deleting the SRX1 gene) renders mutant cells hypersensitive to oxidation stress elicited by exogenous oxidant (such as hydrogen peroxide) 5. To examine if plant Srx would function intracellularly, the cDNAs of Arabidopsis and rice Srxs were chosen as representatives for complementing Srx deficiency in SRX1 deletion yeast cells. From Figure 2, it is clear that ectopic expression of AtSrx or OsSrx restored the growth of SRX1 deletion yeast cells in the presence of hydrogen peroxide, indicating that plant Srx could complement functional deficiency of Srx in SRX1 deletion yeast cells.
Complementation of functional deficiency of Srx in SRX1 deletion yeast cells by Srx homolog from Arabidopsis and rice. Three DNA constructs, pAtSrx (for expressing Arabidopsis Srx), pOsSrx (for expressing rice Srx) and pYES2 (as vector control), were each transformed into SRX1 deletion yeast strain. Recombinant cells were selected (using uracil free medium and PCR amplification of AtSrx or OsSrx coding sequences ) and were further streaked onto plates containing 2% galactose and 2 mM hydrogen peroxide (H2O2). Functional complementation of yeast Srx deficiency by plant Srx homolog is indicated by restored growth of the recombinant SRX1 deletion cells containing pAtSrx or pOsSrx in the presence of H2O2.
Targeting of AtSrx to chloroplast
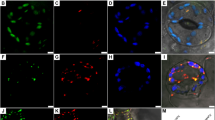
The prediction of chloroplast transit peptide led us to test if plant Srx would be targeted to chloroplast in vivo. To this end, two AtSrx-GFP fusion constructs and one control construct expressing GFP coding sequence alone were transiently expressed in Arabidopsis mesophyll protoplasts, and the patterns of subcellular GFP fluorescence among the three sets of transient expressions were compared using confocal microscopy. In the protoplasts transfected with p35S-AtSrx-GFP, which expressed wild type AtSrx-GFP fusion cistron, the green fluorescence was clearly associated with chloroplasts and colocalized with the red autofluorescence of chloroplasts (Figure 3, top panel). In contrast, in the protoplasts transfected with the control construct p35S-GFP (expressing GFP coding sequence alone, Figure 3, middle panel) or p35S-mAtSrx-GFP (expressing a mutated AtSrx-GFP fusion cistron in which the coding sequence for the predicted chloroplast transit peptide in AtSrx protein was deleted, Figure 3, bottom panel), the green fluorescence was distributed in the cytoplasm surrounding the chloroplasts and was not colocalized with the red autofluorescence of chloroplasts.
Intracellular targeting of AtSrx. Arabidopsis mesophyll protoplasts were transfected with p35S-AtSrx-GFP (expressing a wild type AtSrx-GFP fusion protein, top panel), p35S-GFP (expressing GFP alone, middle panel), or p35S-mAtSrx-GFP (expressing the GFP fusion protein of a truncated AtSrx lacking the first 30 amino acid residues that were predicted to form a chloroplast transit peptide, bottom panel). Expressing protoplasts were examined using dual channel confocal microscopy. Chloroplast targeting is demonstrated by colocalization of GFP fluorescence and chloroplast, and presence of chloroplast-associated yellow signal when green (GFP) and red (chloroplast) fluorescences were merged. Lack of chloroplast targeting is indicated by presence of GFP fluorescence in the cytoplasm surrounding chloroplasts, and the absence of chloroplast-associated yellow signal in the green (GFP) and red (chloroplast) fluorescence-merged image. Bar=20 μM.
Transcriptional patterns of AtSrx in Arabidopsis organs and during stress
Using semiquantitative RT-PCR, the relative transcript levels of AtSrx in Arabidopsis vegetative and reproductive organs were evaluated. AtSrx transcript levels were highest in leaves and intermediate in roots, stems and siliques (Figure 4). In contrast, AtSrx transcription was very low in flowers and required at least 35 amplification cycles to be detected (Figure 4). This transcriptional pattern was reproducibly obtained in multiple independent experiments.
Evaluation of relative transcript levels of AtSrx in Arabidopsis root (R), stem (St), leaf (L), flower (F) and silique (Si) organs using semiquantitative RT-PCR. Amplification of tubulin transcripts was employed to normalize cDNA contents and to monitor the kinetics of thermoamplification during PCR. AtSrx transcript levels were highest in leaf, lowest in flower, and intermediate in other organs. Reproducible transcriptional patterns were obtained in independent assays.
The influence of oxidation stress (conferred by 4 mM hydrogen peroxide), osmotic/water deficit stress (mediated by 10% PEG8000), and cold treatment (8 h at 4 °C) on AtSrx transcript levels was also examined. The application of hydrogen peroxide substantially increased the transcript level of AtSrx as well as those of 2-Cys PrxA and 2-CysPrxB (Figure 5A). Osmotic/water deficit and cold treatments also raised AtSrx transcript level considerably (Figure 5B and 5C). The above transcriptional patterns were each tested in multiple experiments and were found to be reproducible in independent assays.
Changes of AtSrx transcript levels in response to oxidation (by H2O2, A), osmotic/water deficit (by PEG8000, B), and cold (by growing seedlings at 4 °C, C) stresses using semiquantitative RT-PCR. Amplification of tubulin transcripts was used for normalizing cDNA contents and monitoring thermoamplification kinetics. Alterations in the transcript levels of 2-Cys Prx proteins A and B in response to the oxidation stress imposed in this work were also investigated.
Response of AtSrx knock-out line (atsrx1-1) to oxidation stress
Further to above experiments, a T-DNA insertion mutant (atsrx1-1) deficient in AtSrx transcripts was identified (Figure 6A), and its response to oxidation stress was compared to that of wild type Arabidopsis plants in this work. Under standard growth condition, atsrx1-1 and wild type plants were indistinguishable in morphology and development (data not shown). When grown on MS plate containing 10 μM paraquat, a herbicide with a well-known role in increasing ROS production and oxidation stress in chloroplast 31, the growth of atsrx1-1 and that of wild type plants was both affected (Figure 6B). However, the leaf chlorosis phenotype displayed by atsrx1-1 mutant was much more severe than that exhibited by wild type strain (Figure 6B). atsrx1-1 and wild type plants did not differ significantly in Fv/Fm ratio before paraquat treatment (BPT, Figure 6C). However, the decrease of Fv/Fm ratio in the knock-out line (atsrx1-1) was significantly greater than that shown by wild type strain at either 24 h or 48 h after paraquat treatment (APT, Figure 6C). In complementation experiment, the phenotype of atsrx1-1-COMP plants, into which AtSrx cDNA had been reintroduced through Agrobacterium mediated transformation, was similar to that of wild type plants on paraquat containing medium (Figure 6D), indicating that the hypersensitive phenotype of atsrx1-1 plants to paraquat treatment could be corrected by restored expression of AtSrx.
Characterization of AtSrx knock-out mutant (atsrx1-1) and its response to oxidation stress elicited by the herbicide paraquat. (A) A diagram showing the position of T-DNA insertion within the genomic open reading frame of AtSrx (top panel). AtSrx transcripts were detected in wild type (WT) but not atsrx1-1 plants (bottom panel). (B) When grown on MS medium with 10 μM paraquat, the chlorosis phenotype of atsrx1-1 was much more severe than that of WT plant. (C) atsrx1-1 and wild type (WT) plants were similar in Fv/Fm ratio before paraquat treatment (BPT, white column). But at either 24 h (striped column) or 48 h (dotted column) after paraquat (100 μM) treatment (APT), the Fv/Fm ratio was more drastically decreased in atsrx1-1 mutant than in WT plants. (D) In complementation experiment conducted using MS medium containing 10 μM paraquat, the phenotype of atsrx1-1-COMP T1 plants, in which AtSrx expression was restored by genetic transformation was, similar to that of WT plants. By contrast, the atsrx1-1 plants displayed severe chlorosis in their leaves.
Discussion
In this work, four complementary lines of evidence were obtained for the existence of functional Srx protein in both monocot and dicot plant species. First, the deduced amino acid sequences of putative Srxs from eight plant species exhibited significant identities to those of yeast and mammalian Srx proteins, and contained the conserved signature sequence and residues essential for catalysis. Second, ectopic expression of Arabidopsis or rice Srx coding sequence complemented Srx functional deficiency in SRX1 deletion yeast cells. Third, the transcript level of AtSrx was substantially increased under oxidation stress imposed by hydrogen peroxide. Fourth, the knock-out line of AtSrx, atsrx1-1, was more susceptible to oxidation stress elicited by paraquat than wild type Arabidopsis plant. The strong conservation in the primary structure of plant, yeast and human Srxs indicates that there may be high similarity in the basic biochemical mechanism underlying reduction of protein sulfinic acid among eukaryotic organisms. Consequently, the knowledge on yeast and animal Srxs may aid biochemical and molecular analysis of plant Srxs in the future.
Because of the essential role of chloroplast in carbon assimilation and synthesis of amino acids, fatty acids and nucleotides, the redox state of chloroplast must be correctly maintained. In addition to CuZn-superoxide dismutase and ascorbate peroxidase that are known to play a prominent role in detoxifying ROS in chloroplast 32, 33, more recent investigations have shown that 2-Cys Prx proteins are also essential in peroxide removal during photosynthetic electron flux 11. In this work, we have shown that an AtSrx-GFP fusion protein was targeted to chloroplast and that the transcript level of AtSrx was highest in photosynthetic leaves. AtSrx transcription could be substantially increased by oxidant and this increase paralleled with enhanced transcription of 2-Cys Prx proteins. Most importantly, based on the severity of the chlorosis phenotype and the changes in Fv/Fm ratio, which is a sensitive and reliable indicator of stress 26, 27, after paraquat treatment, knock-out of AtSrx expression rendered Arabidopsis plants much more susceptible (than the wild type control) to increased ROS production in chloroplast and this hypersensitivity was reversed by restored AtSrx expression through genetic transformation. Collectively, these data indicate that the expression and function of AtSrx in chloroplast is essential for proper plant response to oxidation stress conditions. Considering that 2-Cys Prx is an important component of antioxidant defense in chloroplast and there exists functional cooperation between 2-Cys Prx and Srx in yeast and mammalian cells 5, 6, 12, it is tempting to suggest that there may be functional cooperation between Srx and 2-Cys Prx in detoxifying peroxides in chloroplast. In this respect, it is interesting to note that 2-Cys Prx with its catalytic cysteine residue modified to sulfinic acid has been detected in chloroplast 34. However, further experiments are needed to examine if plant Srx would indeed interact with 2-Cys Prx and whether plant Srx can reduce Cys-SO2H in vitro and in vivo. It will also be interesting to investigate in more depth if Srx (alone or together with 2-Cys Prx or other partners) would confer protection to chloroplast metabolism including photosynthesis and the synthesis of amino acids, fatty acids and nucleotides.
Because of their sessile life style, plants have evolved sophisticated mechanisms to respond to environmental stresses. A general feature of plant response to biotic and abiotic stresses is production of ROS 10. Although stress generated ROS have recently been proposed to act as signaling molecules to facilitate plant adaptation to adverse environments 35, the underlying mechanism(s) is still not well understood. In yeast and mammalian cells, reversible oxidation of 2-Cys Prx, which involves the function of Srx, has been found essential in mediating hydrogen peroxide-induced gene expression changes 12, 36. Interestingly, drought and cold stresses have been shown to increase the amount of 2-Cys Prx with its catalytic cysteine residue modified to Cys-SO2H in barley 37, though it is still unknown if there would be any changes in the reduction of this over-oxidized 2-Cys Prx during stress. We have demonstrated in this work that osmotic/water deficit and cold stresses could substantially increase the transcript level of AtSrx, indicating that there may be enhanced reduction of protein sulfinic acid by Srx under these stress conditions. In line with this reasoning, signaling by ROS has been considered to be an integral part of plant responses to osmotic/water deficit and cold stresses 38. The next challenges are to understand potential impact of enhanced Srx expression on reversible oxidation of 2-Cys Prx and its implication on ROS mediated signaling in plants under stress.
From above discussions, it might be understandable why Srx was found to be highly expressed in leaves and its transcripts were further increased under stresses. However, AtSrx transcripts were also detected in roots, flowers and siliques albeit at relatively lower levels. This suggests that the physiological function of Srx may be required in most plant tissues under normal growth condition, but its expression level in different tissues (and organs) is subject to transcriptional regulation. A recent study has revealed that the level of protein oxidation is drastically reduced during bolting and flowering in Arabidopsis 39. This provides an important clue for further investigations into potential mechanism involved in transcriptional regulation of AtSrx during flower development.
In summary, this work has generated novel insights into the existence and potential functionality of Srx homolog in higher plants. Based on this and previous investigations, it is hypothesized that there may be functional interaction between Srx and 2-Cys Prx proteins in plant cells. This interaction is likely to be important for maintaining redox balance in chloroplast and enabling plants to adapt to abiotic environmental stresses that involve ROS production. To advance this work, we are now in the process to analyze the biochemical mechanism and biological function of Srx in plant cells.
References
Giles NM, Watts AB, Giles GI, et al. Metal and redox modulation of cysteine protein function. Chem Biol 2003; 10:677–693.
Jacob C, Giles GI, Giles NM, Sies H . Sulfur and selenium: The role of oxidation state in protein structure and function. Angew Chem Int Ed Engl 2003; 42:4742–4758.
Jacob C, Holme AL, Fry FH . The sulfinic acid switch in proteins. Org Biomol Chem 2004; 2:1953–1956.
Buchanan BB, Balmer Y . Redox regulation: A broadening horizon. Annu Rev Plant Biol 2005; 56:187–220.
Biteau B, Labarre J, Toledano MB . ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 2003; 425:980–984.
Chang TS, Jeong W, Woo HA, et al. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J Biol Chem 2004; 279:50994–51001.
Woo HA, Jeong W, Chang TS, et al. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem 2005; 280:3125–3128.
Wood ZA, Schroder E, Robin-Harris J, Poole LB . Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 2003; 28:32–40.
Jonsson TJ, Murray MS, Johnson LC, Poole LB, Lowther WT . Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin. Biochemistry 2005; 44:8634–8642.
Apel K, Hirt H . Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 2004; 55:373–399.
Konig J, Baier M, Horling F, et al. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci USA 2002; 99:5738–5743.
Bozonet SM, Findlay VJ, Day AM, et al. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J Biol Chem 2005; 280:23319–23327.
Wiegel D, Glazebrook J . Arabidopsis: A Laboratory Manual. New York: CHSL Press, 2002.
Sambrook J, Russell DW . Molecular Cloning: A Laboratory Manual. New York: CHSL Press, 2001.
Li D, Zhu H, Liu K, et al. Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 2002; 277:27772–27781.
Gietz RD, Woods RA . Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 2002; 350:87–96.
Ling M, Merante F, Robinson BH . A rapid and reliable DNA preparation method for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res 1995; 23:4924–4925.
Damm B, Schmidt R, Willmitzer L . Efficient transformation of Arabidopsis thaliana using direct gene transfer to protoplasts. Mol Gen Genet 1989; 217:6–12.
Abel S, Theologis A . Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 1994; 5:421–427.
Bhushan S, Lefebvre B, Stahl A, et al. Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep 2003; 4:1073–1078.
Li G, Liu K, Baldwin SA, Wang D . Equilibrative nucleoside transporters of Arabidopsis thaliana cDNA cloning, expression pattern, and analysis of transport activities. J Biol Chem 2003; 278:35732–35742.
del Pozo JC, Allona I, Rubio V, et al. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 1999; 19:579–589.
Horling F, Lamkemeyer P, Konig J, et al. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol 2003; 131:317–325.
van der Weele CM, Spollen WG, Sharp RE, Baskin TI . Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J Exp Bot 2000; 51:1555–1562.
Sakamoto H, Maruyama K, Sakuma Y, et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 2004; 136:2734–2746.
Kraus GH, Weis E . Chlorophyll fluorescence and photosynthesis: The basis. Annu Rev Plant Physiol 1991; 136:472–479.
Schreiber U, Bilger W, Neubauer C . Chlorophyll fluorescence as a non- intrusive indicator for rapid assessment of in vivo photosynthesis. In: Schulze ED, Caldwell MM. eds. Ecophysiology of Photosynthesis (Ecological Studies). Springer: Berlin Heidelberg New York 1994:49–70.
Romero HM, Berlett BS, Jensen PJ, Pell EJ, Tien M . Investigations into the role of the plastidial peptide methionine sulfoxide reductase in response to oxidative stress in Arabidopsis. Plant Physiol 2004; 136:3784–3794.
van Engelen FA, Molthoff JW, Conner AJ, et al. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 1995; 4:288–290.
Bechtold N, Ellis J, Pelletier G . In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Pairs, Life Sciences 1993; 316:1194–1199.
Kunert KJ, Dodge AD . Herbicide-induced radical damage and antioxidative systems. In: Boger P, Sandmann G. eds. Target Sites of Herbicide Action. Boca Raton: CRC Press, 1989:45–63.
Schurmann P . Redox signaling in the chloroplast: the ferredoxin/thioredoxin system. Antioxid Redox Signal 2003; 5:69–78.
Baier M, Dietz KJ . Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J Exp Bot 2005; 56:1449–1462.
Dietz KJ . Plant peroxiredoxins. Annu Rev Plant Biol 2003; 54:93–107.
Foyer CH, Noctor G . Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 2005; 17:1866–1875.
Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA . Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol 2005; 17:183–189.
Konig J, Lotte K, Plessow R, et al. Reaction mechanism of plant 2-Cys peroxiredoxin. Role of the C terminus and the quaternary structure. J Biol Chem 2003; 278:24409–24420.
Xiong L, Schumaker KS, Zhu JK . Cell signaling during cold, drought, and salt stress. Plant Cell 2002; 14 Suppl:S165–S183.
Johansson E, Olsson O, Nystrom T . Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem 2004; 279:22204–22208.
Acknowledgements
We thank Professors Karl-Josef Dietz (Department of Biochemistry and Physiology of Plants W5, University of Bielefeld, Germany) and Hong Jie Li (Institute of Crop Science, Chinese Academy of Argicultural Sciences, China) for constructive comments on this work and the manuscript. This work was supported by a grant (KSCX2-SW-304) from the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, X., Liu, X., Zhang, J. et al. Molecular and functional characterization of sulfiredoxin homologs from higher plants. Cell Res 16, 287–296 (2006). https://doi.org/10.1038/sj.cr.7310036
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7310036
Keywords
This article is cited by
-
Antioxidant enzymes that target hydrogen peroxide are conserved across the animal kingdom, from sponges to mammals
Scientific Reports (2023)
-
Identification of genomic regions governing moisture and heat stress tolerance employing association mapping in rice (Oryza sativa L.)
Molecular Biology Reports (2023)
-
Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica
Scientific Reports (2016)
-
AtTHIC, a gene involved in thiamine biosynthesis in Arabidopsis thaliana
Cell Research (2008)
-
Protein cysteine sulfinic acid reductase (sulfiredoxin) as a regulator of cell proliferation and drug response
Oncogene (2008)