ABSTRACT
Using two-colour flow cytometry >200 antibodies submitted to the 8th International Workshop of Human Leukocyte Differentiation Antigens (HLDA8) have been analyzed for their reactivity with resting and activated CD203c+ basophils. Four antibodies either non-reactive or weakly reactive with resting basophils exhibited an increased reactivity with basophils activated by anti-IgE-mediated cross-linking of the high affinity IgE receptor (FcεRI). These include antibodies against CD164 (WS-80160, clone N6B6 and WS-80162, clone 67D2), as well as two reagents with previously unknown specificities that were identified as CD13 (WS-80274, clone A8) and CD107a (WS-80280, clone E63-880). The activation patterns followed either the “CD203c-like” or “CD63-like” activation profile. The CD203c profile is characterized by a rapid and significant upregulation (of CD13, CD164, and CD203c), reaching maximum levels after 5-15 min of stimulation. The phosphoinositide-3-kinase (PI3K)-specific inhibitor wortmannin inhibited the upregulation of these markers whereas 12-O-tetradecanoyl-phorbol-13-acetate (TPA) induced a rapid and FcεRI-independent upregulation within 1-2 min. In the CD63 profile, maximum upregulation (of CD63 and CD107a) was detected only after 20-40 min, and upregulation by TPA reached maximum levels after 60 min. In summary, our data identify CD13, CD107a, and CD164 as novel basophil-activation antigens. Based on time kinetics of upregulation, we hypothesize that molecules of the “CD203c group” and the “CD63 group” are linked to two different mechanisms of basophil activation.
Similar content being viewed by others
INTRODUCTION
Basophils and mast cells are important effector cells of inflammatory reactions 1, 2, 3. In contrast to eosinophils and neutrophils, they possess high-affinity immunoglobulin (Ig) E receptors (FcεRI) that are cross-linked upon engagement of receptor-bound IgE with allergens, resulting in the release of several mediators to the extracellular space or in the transport of vesicle-bound membrane proteins to the plasma membrane 4, 5. Although the cell surface marker profile of basophils parallels that of mast cells to a degree, basophils are identified by their strong expression of interleukin (IL)-3Rα (CD123) but not of c-kit (CD117), whereas mast cells express high levels of CD117 but not CD123 6, 7, 8, 9.
Basophils and mast cells originate in the bone marrow from CD34+ progenitor cells that coexpress the differentiation and activation antigen ecto-nucleotide pyrophosphatase/phosphodiesterase 3 (CD203c) 10, 11. This antigen was identified as a selective cell surface marker for the basophil and mast cell lineage 10, 11. Apart from CD203c, a number of other basophil-specific markers have been described which are expressed either on the cell surface (Bsp-1) 12, 13 or in cytoplasmic compartments (2D7, BB1) 14, 15, 16 of mature basophils. However, none of these basophil-specific markers are molecularly identified and no information exists about their expression in early basophil and/or mast cell differentiation.
Basophils challenged by allergens via the FcɛRI secrete several mediators including histamine and leukotriene C4 17, 18. In addition, activation markers like the granule-associated molecule CD63 and the ecto-enzyme CD203c are upregulated to the plasma membrane in response to FcɛRI cross linkage 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30. These markers are now routinely used for flow cytometry-based basophil activation tests. However, the population size of activated basophils as defined by the CD63 and CD203c tests differs significantly. Thus, only a subpopulation of basophils that is sensitive to allergen-induced CD203c upregulation shows also an upregulated expression of CD63 31. Moreover, the kinetic profile of CD63 upregulation and the sensitivity to inhibitors and activators of FcɛRI-mediated signaling differs from that of CD203c 31. In a recent report it was demonstrated that tetradecanoyl phorbol acetate (TPA), a stimulator of the protein kinase C (PKC) pathway, accelerated and enhanced CD203c upregulation, but delayed CD63 upregulation 31. In contrast, the phosphoinositol-3 kinase (PI-3) inhibitor wortmannin effectively inhibited CD203c as well as the CD63 upregulation 31, 32.
Several antibodies with known and unknown specificity for cell surface antigens were submitted to the HLDA8 conference and workshop 2004. Of these, > 260 antibodies were selected to be analyzed for their reactivity with resting and activated basophils. The aim of the current work was to identify potential novel and selective markers for basophils and their activated counterparts. To address this question, resting and FcɛRI-activated basophils were analyzed for their coexpression of CD203c and the antigens defined by workshop antibodies. Based on results obtained from these analyses, we provide evidence that CD13, CD107a (and to a minor extent CD107b), and CD164 are novel basophil activation antigens that follow different upregulation profiles.
MATERIALS AND METHODS
Individuals
The peripheral blood of healthy individuals was drawn after informed consent was given. Lung tissue was obtained from surgical specimens obtained from patients with bronchiogenic carcinoma (n=2). Informed consent was obtained before surgery. Tissue and blood samples were obtained and prepared following the guidelines of the local ethics committees. MC were isolated according to published techniques 33, 34. In brief, lung tissue was cut into small pieces and then washed extensively in Tyrode's buffer. Tissue-fragments were incubated 2 times in collagenase type II ( 2 mg/ml) for 60 min at 37°C. After digestion, cells were recovered, washed, and examined for the presence and percentage of MC by Wright Giemsa staining. Isolated MC were cultured in RPMI 1640 medium and 10% fetal calf serum (FCS) at 37°C for at least 12 h before use in immunostaining experiments.
Flow cytometry-based basophil activation test
Heparinized peripheral blood (PB) cells (90 μl) were stimulated with either 10 μl (1 μg/ml final concentration) goat anti-human IgE antibody (ICN Biomedicals, Irivine CA, USA) or phosphate-buffered saline (PBS) for 15 min at 37°C, as previously described 19. In selected experiments stimulation times were modified (range: 2-60 min). To stop the Ca2+-dependent reaction, cells were washed in PBS/20mM EDTA and resuspended in 50 μl FACS buffer (0.1% NaN3 + 0.1% BSA in Hank's balanced salt solution). The cells were then stained by the following procedure: In a first step, cells were incubated with 10 μl of the indicated workshop mAb for 15 min on ice. After washing in FACS buffer, the cells were stained with the (Fab)′2 fragment of a fluorescein isothiocyanate (FITC)-coupled rabbit anti-mouse antiserum (DakoCytomation, Glostrup, Denmark) for 15 min on ice. After washing in FACS buffer, the cells were incubated for 15 min on ice with a non-binding mouse IgG antibody to block free binding sites of the secondary-step reagent (Southern Biotech). Finally, the cells were stained with 10 μl (1 μg/ml final concentration) of the phycoerythrin (PE)-coupled CD203c-specific antibody 97A6 (Immunotech, Marseille, France) for 15 min on ice. In some experiments, the cells were double-stained either with 97A6-PE and anti-CD63-FITC (Becton Dickinson, Heidelberg, Germany), anti-CD107a-FITC, anti-CD107b-FITC (kind gifts of Dr. Enoc Hollemweguer, PharMingen, San Diego, CA), or anti-CD13-FITC (Immunotech). After red blood cell lysis with 100 μl Optilyse lysing reagent (Immunotech), the cells were washed, resuspended in FACS buffer, and analyzed on a FACSCalibur flow cytometer (Becton Dickinson). Modulation of antigen expression was evaluated by calculating the stimulation index (SI) as previously described 19.
For blocking/activation experiments, 500 nM Wortmannin (Sigma-Aldrich, Steinheim, Germany), 500 nM TPA (Sigma-Aldrich), or 10 μM prostaglandin D2 (Sigma-Aldrich) were used. In the case of wortmannin, PB cells were pre-incubated with the inhibitor for 15 min at 37°C prior to stimulation with anti-IgE antibody. In other experiments, PB cells were stimulated with prostaglandin D2 or TPA for various time intervals.
Identification of antibody-defined antigens
The specificities of the submitted workshop antibodies 80280 (E63-880) and 80274 (A8) were not known. To identify the detected molecules, lysates from the basophil cell line KU-812 were immuno-precipitated with the respective antibodies. 5×107 KU-812 cells were lysed in 0.5 ml RIPA buffer containing 1.5 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 4 mM EDTA and PBS, supplemented with 4 ìg/ml aprotinin, and 1 mM PMSF (lysis buffer). The immunoprecipitations were performed in lysis buffer using Protein G Sepharose 4 fast flow (Amersham Bioscienses, Uppsala, Sweden) according to the protocol of the supplier. Either 7μg of purified A8 antibody, 100 μl of E63-880 antibody, or unspecific control IgG was used. The samples were separated on 10% SDS-PAGE, stained with Coomassie dye, and bands that were specifically precipitated with E63-880 or A8 antibodies but not with control antibody were cut and subjected to nanoHPLC-ESI-MS/MS analysis.
NanoHPLC-ESI-MS/MS
The interested gel bands were digested using trypsin (sequencing grade, Promega, Mannheim, Germany) and then analyzed with a Dionex LC Packings HPLC system (Dionex LC Packings, Idstein, Germany) containing the components FAMOS (autosampler), Switchos (loading pump and switching valves), and Ultimate (separation pump and UV detector). The ESI-MS/MS spectra were recorded using a high performance quadrupole Time-of-Flight (QTOF) mass spectrometer QStar (Applied Biosystems, Applera, Darmstadt, Germany), equipped with a nano-ESI source (column adapter and distal coated SilicaTips (New Objective, Woburn, USA)). The ESI-MS/MS-spectra were correlated with the NCBInr-protein sequence database using Mascot software, available online at http://www.matrixscience.com.
RESULTS
Reactivity of HLDA8 antibodies with basophils, mast cells, HMC-1 and KU-812 cells
A total of 262 workshop monoclonal antibodies (mAb) were tested for their reactivity with primary peripheral blood (PB) basophils, mast cells from lung tissues, as well as cells of the mast cell line HMC-1 and the basophil cell line KU-812. Tab. 1 summarizes the most relevant mAbs that were positive for either basophils or mast cells (Δ MFI > 2.5). Several markers that reacted with primary basophils but were negative on lung mast cells were identified (shaded background). In 11/17 cases, these markers were also found on KU-812 cells. In contrast, they were all negative on HMC-1 cells. The basophil-selective markers include CD62L, TRAIL R1 and R2, CCR1, CCR2, and CCR5, CXCR1, CD123 (IL-3Rα), IL-18Rα, CD244, Siglec-9, BSP-1, TLR-4, OX108, FGFR2, and FGFR3. All markers that were positive on mast cells were also expressed on basophils. These data show the identification of a variety of markers selective for basophils but not mast cells. However, none of the markers were selective for mast cells.
Reactivity of HLDA8 antibodies with resting and activated basophils
The workshop mAbs were also tested for their reactivity with resting and activated basophils. Resting basophils were defined as CD203c+ peripheral blood (PB) cells incubated in PBS, whereas activated basophils were obtained by stimulation with anti-IgE antibody and were characterized by upregulated CD203c cell surface expression (CD203c++). Tab. 2 summarizes selected antigens expressed on resting and/or activated basophils, including CD62L, anti-Rh 17, CXCR1, CXCR4, CCR1, CCR2, CD164, CD123, IL-18Rα, BSP-1, OX108, and LAIR. Three of these antigens were either upregulated (CD164, unknown 80274 antigen) or de novo expressed (unknown 80280 antigen) on activated basophils (Δ MFI > 2.5). In contrast, the BSP-1 antigen (80431) was downregulated on activated basophils to levels near the control value.
Identification of novel activation markers
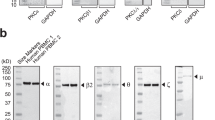
Tab. 2 revealed that the antigens detected by mAbs 80162 (CD164), 80274 (unknown), and 80280 (unknown) were >2-fold upregulated on activated basophils. To identify the unknown specificity of mAbs 80274 and 80280, KU-812 lysates were immuno-precipitated with these mAbs, the precipitated material was then separated by SDS-PAGE, and the bands not appearing in control precipitates were cut und subjected to nanoHPLC-MS/MS analysis. The analysis of the 170 kD bands shown in Fig. 1 revealed that mAb 80274 recognizes aminopeptidase N (CD13) 35 and mAb 80280 lysosomal-associated membrane glycoprotein-1 (LAMP-1; CD107a) 36, 37, 38, 39. The specificity of these antibodies was further confirmed by their selective recognition of BW cells transfected with human CD13 (personal communication of Dr. H. Stockinger, Vienna, Austria) and of CHO cells transfected with human CD107a, respectively (personal communication of Dr. P. Meikle, Adelaide, Australia).
Identification of A8- and E-63-880 antigens. Unknown antigens detected by antibodies A8 (80274) and E63-880 (80280) were immunoprecipitated from KU-812 cells with protein G sepharose and identified by nanoHPLC-ESI-MS/MS analysis. (A) Antibody A8 detected a band with apparent molecular mass of 170kD that was identified as CD13. (B) Antibody E63-880 precipitated a band of similar size that was identified as CD107a (although the published molecular mass of CD107a is 120 kD 37, some myeloid cells express highly glycosylated forms of CD107a with the apparent molecular mass of >160 kD 54).
Upregulation profiles of basophil activation markers
The profiles of FcεRI-mediated upregulation of antigens detected by workshop antibodies (CD13, CD107a, and CD164) and of the known activation markers CD63 and CD203c were compared. In addition, CD107b (LAMP-2) and the basophil-specific antibody BSP-1 were studied. CD107b was selected because it shares structural similarity and lysosomal expression with CD107a, and is upregulated in activated mast cells in a similar fashion as CD107a 37. BSP-1 was of interest because it is the first reported cell surface marker that is highly specific for basophils 12, 13. For these experiments, resting and activated basophils were stained with the selected FITC-conjugated antibodies and counter-stained with the CD203c-specific antibody conjugate 97A6-PE. The plots in Fig. 2 demonstrate that CD107a (A) and CD107b (B) are not expressed on the surface of resting basophils. However, a strong upregulation of CD107a is observed in a supopulation of activated basophils. In contrast, CD107b is only weakly upregulated. The profile of CD107a and CD107b parallels that of CD63 upregulation (C), indicating that CD63, CD107a, and CD107b are localized in the same intracellular compartment and are expressed by the same subset of activated basophils. Indeed, coexpression analysis of CD63 versus CD107a confirmed that these molecules are upregulated in the same basophil subpopulation (not shown). In contrast to the other molecules, CD13 (D) and CD164 (E) are upregulated on almost all basophils and their profiles parallel that of CD203c. Unlike the other markers, the BSP-1 antigen was not upregulated but it almost completely disappeared after basophil activation (F). Most likely, the antigen does not irreversibly disappear from the surface but is rather endocytosed and re-distributed to the cell surface, because maintained stimulation (>40 min) leads to a reappearance of BSP-1 antigen (not shown).
Differential expression of markers on resting and activated basophils. Heparinized PB cells were incubated at 37°C either with 1μg/ml of an activating anti-IgE antibody (right panel) or with PBS (left panel). Reactions were stopped after 20 min by adding 20mM EDTA. The cells were labeled with the indicated workshop antibodies and stained with a FITC-conjugated anti-mouse IgG antibody. After blocking free binding sites with excess mouse IgG, the cells were stained with a PE-conjugated CD203c-specific antibody (97A6), that is specific for basophils. CD107b and CD63 were stained by direct immunofluorescence using a FITC-conjugate. 50,000 cells were analyzed on a FACSCalibur flow cytometer using the CellQuest software.
Differential expression of CD164 epitopes on resting and activated basophils
CD164 is a heavily glycosylated sialomucin that contains three antibody-defined epitopes which are grouped into sialidase-sensitive (group I), N-glycosidase-sensitive (group II), and glycosidase-resistant (class II) epitopes 40, 41, 42, 43. To analyze CD164 epitope expression on basophils, PB cells were stained with the class III epitope-reactive antibodies 67D2 (80162) and N6B6 (80160), the class II reactive antibody 103B2, and the class I reactive antibody 105A5. As shown in Fig. 3, the class I epitope is not expressed on any basophil subset, the class II epitope is weakly expressed only on activated basophils, whereas all basophils are positive for the class III epitope of CD164. However, the expression of this epitope is weak on resting basophils but 11 – 18-fold upregulated on activated basophils. This demonstrates that CD164 epitopes are not only differentially expressed on stem cells but also on basophils.
Differential expression of CD164 epitopes on basophils. PB basophils were analyzed for their expression of different CD164 epitopes. Antibodies N6B6 and 67D2 define class III, antibody 103B2 class II, and antibody 105A5 class I epitopes. Resting (left panel) and activated cells (right panel) were stained and analyzed as described in Fig. 2. Values in the plots represent median fluorescent intensities of CD164 signals within the CD203c+ populations.
Kinetic profiles of basophil activation marker upregulation
To study the time course of activation marker upregulation, PB cells were stimulated with anti-IgE antibody for 2 – 120 min prior to staining. Fig. 4A shows that CD203c, CD164, and CD13 reached the half-maximum upregulation already after 2 min and the plateau value after 10 – 20 min. In contrast, CD63 and CD107a upregulation was delayed, the half-maximum value was observed only after about 10 min, and complete upregulation was reached after 25 – 30 min. Moreover, CD107a and CD63 upregulation was detected only in a subset of activated basophils, whereas CD13, CD164, and CD203c upregulation was observed in virtually all basophils.
(A) Kinetic profiles of basophil activation marker upregulation. Heparinized PB cells were stimulated with 1μg/ml of a cross-linking anti- IgE antibody. The activation was stopped after 2, 5 and 25 min with 20mM EDTA. Cells were then stained and analyzed as described in Fig. 2. The percentage of upregulated CD203c+ cells was defined as activated basophils and plotted versus the time of stimulation. (B) Effect of TPA on activation marker upregulation. PB cells were stimulated with 500nM TPA for 2, 10, 20 and 60 min. After stopping the reaction with 20mM EDTA, the cells were stained and analyzed as described in Fig. 2.
Fig. 4B shows that the PKC stimulator TPA induced a rapid upregulation of CD203c, CD13, and CD164 on basophils. Notably, TPA-induced CD203c surface expression was observed even earlier than anti-IgE-induced CD203c upregulation. In contrast, the onset of CD63 and CD107a upregulation was markedly delayed compared to anti-IgE-induced upregulation. Indeed, half-maximum upregulation was reached only after 25 – 30 min. These data suggest that the kinetic profiles of activation molecules can be divided into a “fast responder” (CD203c group) and into a “slow responder” group (CD63 group). Members of the fast responders are CD203c, CD164, and CD13, whereas slow responders consist of CD63, CD107a (and most likely CD107b).
Effect of wortmannin and prostaglandin D2 on upregulation of activation markers
To analyze whether the upregulation of activation markers is dependent on the activity of the signaling enzyme PI-3K, PB cells were pre-incubated with the PI-3K inhibitor wortmannin prior to stimulation with anti-IgE antibody. Again, Fig. 5A demonstrates that the activation marker profiles can be divided into group CD203c and group CD63 molecules. Members of the CD203c group showed a half-maximum reduction of marker upregulation on PB cells preincubated with 500 nM wortmannin. In contrast, the upregulation of CD63 and CD107a was almost completely inhibited at this wortmannin concentration.
(A) Effect of wortmannin on anti-IgE-induced activation marker upregulation. Heparinized PB cells were incubated with 500nM wortmannin for 15 min at 37°C prior to stimulation with 1 μg/ml anti-IgE antibody. After stopping the reaction, cells were stained and analyzed as described in Fig. 2 (B) Effect of prostaglandin D2 (PGD2) on activation marker upregulation. PB cells were stimulated with 10 μM PGD2 for 10 min at 37°C. After stopping the reaction, cells were stained and analyzed as described in Fig. 2.
The receptor for prostaglandin D2 (or CRTH2) is a molecule expressed on TH2 cells, eosinophils and basophils 44. Recently, it was reported that the cognate ligand, prostaglandin D2 (PGD2), potently activates the upregulation of CD203c but not CD63 45. In these studies, we investigated whether the studied activation markers follow one of the described kinetic behaviors. As shown in Fig. 5B, PGD2 not only induces the upregulation of CD203c but also the other members of this group, CD13 and CD164. Also, and in line with the published data, no CD63 and no CD107a upregulation could be observed, even after one hour of stimulation with PGD2. These data provide further evidence of the existence of two basophil activation pathways.
DISCUSSION
Basophils and mast cells share many common functional and phenotypic features. In the bone marrow, basophils and basophil progenitors coexist with mast cells and their precursors. It is therefore a major challenge to identify suitable markers to distinguish mast cells from basophils. Several reports have shown that the α-chain of the IL-3 receptor (CD123) is a key membrane molecule expressed on basophils but not on mast cells, and vice versa, the receptor tyrosine kinase c-kit (CD117) is exclusively found on mast cells but not on basophils 4. Employing a panel of > 260 antibodies submitted to the 8th HLDA workshop conference, we describe here the identification of novel surface markers that are able to distinguish basophils from mast cells. In particular, we show that CD62L, TRAIL-R1 and TRAIL-R2, CCR1, CCR2, and CCR5, CXCR1, CD123 (IL-3Rα), IL-18Rα, CD244, Siglec-9, BSP-1, TLR-4, OX108, FGFR2, and FGFR3 are basophil markers that are not expressed on mast cells. As most of these molecules are receptors, it is likely that basophils interact with different ligands than mast cells, and that basophil activation is regulated by other mechanisms than mast cell activation.
Since more than a decade, flow cytometry-based assays have been developed to distinguish resting from activated basophils and to monitor activation of basophils. The most prominent markers are the tetraspanin CD63 46 and the ecto-enzyme CD203c 47, 48, 49. Both markers are suitable to analyze allergen-induced basophil activation and are routinely used in allergy diagnosis 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30. In the search for additional markers, we analyzed a panel of workshop antibodies for differential reactivity with resting and activated basophils. In this screen, three new activation antigens were identified that were upregulated on basophils after challenge with anti-IgE antibody, including CD164 40, 41, 42, 43 and two antibody-defined antigens of unknown specificity. The unknown antigens were identified as CD13 and CD107a by nanoHPLC-ESI-MS/MS analysis and by the specific reactivity of the respective antibodies with transfectant lines expressing CD13 35 and CD107a 36, 37, 38, 39.
CD63 was originally identified in the granules of platelets and described as a platelet activation marker, but later, a predominant expression of CD63 was also found in the granule membranes of basophils 46. Activation-induced cross-linking of the IgE receptor by allergen or anti-IgE antibody leads to the activation of downstream molecules and migration and fusion of the granules to the plasma membrane. As a result, histamine and other mediators are released to the extracellular space and CD63 is translocated to the plasma membrane. Electron microscopic studies revealed that the time course of histamine release and CD63 upregulation correlates with the anaphylactic degranulation (AND) of large granules but not with piecemeal degranulation (PMD) of small vesicles 17. As CD203c upregulation follows a different time course, it is likely that this pathway is not associated with AND. According to the time course of upregulation on basophils, the antigens could be divided into a CD63-like and a CD203c-like group. Members of the CD63 group include CD63 and CD107a, whereas CD203c, CD13, and CD164 belong to the other group. Several lines of evidence indicate that the translocation of the described activation markers from cytoplasmic compartments to the extracellular space follow different activation pathways. First, the kinetic profiles of CD107a and CD63 upregulation differ significantly from that of CD13, CD164, and CD203c. In fact, the maximum FcɛRI-mediated upregulation of group CD63 molecules required a 3-fold longer activation time than that of CD203c, CD13, or CD164. Second, inhibitors and activating molecules exhibited the same effects on CD63 and CD107a upregulation but acted in a different manner on group CD203c molecules. Thus, stimulation with TPA enhanced the upregulation of CD203c, CD13, and CD164 but delayed the upregulation of group CD63 molecules, when compared to FcɛRI-mediated stimulation. In addition, the inhibitory effect of wortmannin was much more pronounced for group CD63 molecules than for group CD203c molecules. Moreover, IL-3 and prostaglandin D2 induced the upregulation of CD203c, but not the upregulation of CD63. These observations suggest that CD63 and CD107 are stored in the same granules, whereas group CD203c molecules are stored in separate vesicles. As shown in Fig. 2, activated basophils consist of CD203c+CD63/CD107a− and CD203c+CD63/CD107a+ cells. The fact that the CD63+ and the CD107a+ populations are identical (as revealed by coexpression analysis) further supports the view that the signaling cascade involved in the upregulation of CD63 may be distinct from that required for CD203c upregulation. In this context it is tempting to speculate that molecules of the CD63 group are located in AND-associated granules, whereas CD203c group molecules are stored in different compartments. To address this question, studies are under way to analyze the cellular distribution of the described activation markers in detail.
A striking feature of all members of the CD203c group is that they are not only markers of activated but also of resting basophils. In this context it is noteworthy that two of these molecules, CD13 and CD203c, are ecto-enzymes, and that an additional ecto-enzyme, CD26 50, follows a similar upregulation kinetic profile as CD13. These enzymes are known to cleave nucleotides and their derivatives (CD203c) as well as N-terminal mono- and dipeptides (CD13 and CD26, respectively). It is therefore likely that ecto-enzymes play an important role in the immediate (earliest) events of basophil activation preceding later events like the release of histamine and other granule-associated mediators.
We have shown that CD164 is differentially expressed on resting and activated basophils. In this context it is of interest that only the peptide class III epitopes are weakly expressed on resting basophils, whereas the N-glycosidase-sensitive class II epitopes and the sialidase-sensitive class I epitopes are not expressed. On activated basophils, the class III epitopes are highly upregulated, the class II epitopes are de novo expressed, and the class I epitopes remain absent. The differential expression of CD164 epitopes on basophils resembles the differential expression on hematopoietic stem cells. In particular, CD34+ bone marrow cells express high levels of class III epitopes and low levels of class I epitopes at low selectivity, whereas the class II epitopes are expressed at high levels and with high selectivity in this population 40, 41, 42, 43. The highest expression is found in the CD34+CD38− fraction 41. Thus, CD164 epitopes are not only differentially expressed in early hematopoietic stem cell subsets, but also on resting and activated basophils.
Recently, the structurally related lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2; CD107a and CD107b) have been identified as activation markers in a variety of cells [36-39, 51-53]. Thus, both molecules were reported to be expressed on activated but not resting platelets, and CD107a was shown to be expressed exclusively on cytotoxic NK effector cells but not on other NK cell subsets 53. Moreover, CD107a is a selective marker to further dissect antigen-specific (“tetramer”-positive) T cells into rare antigen-reactive cytotoxic T effector cells and into regulatory T cells or antigen-specific T cells with other functions 52. Very recently, CD107a and CD107b were also identified as reliable activation markers for human mast cells. After stimulation with anti-IgE, a rapid translocation of cytoplasmic CD107 molecules to the cell membrane associated with a release of histamine, leukotriene C4, and prostaglandin D2, was observed 51. In the current study, we describe a new subset of immune effector cells with an activation-induced expression of CD107a and — to a weaker extent — CD107b. As the CD107a-specific antibody 80280 was submitted to the HLDA8 workshop as specific for dendritic cell subsets, it is possible that CD107a is also a marker for activated dendritic cells. Studies to address this question are in progress.
The identification of novel basophil activation markers as well as the analysis of their upregulation profiles and responsiveness to various inhibitory and activating agents revealed the existence of at least two distinct activation pathways. A well established hypothesis is that fusion of basophil granules to the plasma membrane and the resulting release of histamine are not the first events that can be detected in basophil activation. In the present study we show that upregulation of “group CD203c molecules” is an earlier event that can be easily analyzed by flow cytometer-based methods. We further show that most members of this group are ecto-enzymes which may play an important role in the immediate early events of basophil activation. In addition, we present CD107a as a potential universal activation marker of hematopoietic effector cells including platelets, NK cells, antigen-reactive T cells, mast cells, and basophils. Finally, our data support the hypothesis that upregulation of group CD63 molecules is associated with anaphylactic degranulation (AND) whereas group CD203c molecules are upregulated by other pathways.
References
Schwartz LB . The mast cell. In: Kaplan AP, ed. Allergy. Edinburgh, Churchill Livingston 1985; 1:53.
Galli SJ . Biology of disease: New insights into “the riddle of mast cells”: Microenvironmental regulation of mast cell development and phenotypic heterogeneity. Lab Invest 1990; 62:5–33.
Valent P, Bettelheim P. The human basophil. Crit Rev Hematol Oncol 1990; 10:327–52.
Valent P . The phenotype of human eosinophils, basophils, and mast cells. J Allergy Clin Immunol 1994; 94:1177–83.
Falcone FH, Haas H, Gibbs BH . The human basophil: a new appreciation of its role in immune responses. Blood 2000; 96:4028–38.
Valent P, Bettelheim P . Cell surface structures on human basophils and mast cells: Biochemical and functional characterization. Adv Immunol 1992; 52:333–423.
Valent P, Besemer J, Muhm M, et al. Interleukin-3 activates human basophils via high affinity binding sites. Proc Natl Acad Sci U S A 1989; 86:5542–6.
Valent P . The riddle of the mast cell: c-kit ligand as the missing link? Immunol Today 1994; 15:111–4.
Valent P, Besemer J, Sillaber CH, et al. Failure to detect interleukin-3 binding sites on human mast cells. J Immunol 1990; 145:3432–7.
Bühring H-J, Simmons PJ, Pudney M, et al. The monoclonal antibody 97A6 defines a novel surface antigen expressed on human basophils and their multi- and unipotent progenitors. Blood 1999; 94:2343–56.
Bühring HJ, Seiffert M, Giesert C, et al. The basophil activation marker defined by antibody 97A6 is identical with ecto-nucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP3; CD203c). Blood 2001; 97:3303–05.
Bodger MP, Mounsey GL, Nelson J, Fitzgerald PH . A monoclonal antibody reacting with human basophils. Blood 1987; 69:1414–18.
Bodger MP, Newton LA . The purification of human basophils: their immunophenotype and cytochemistry. Br J Haematol 1987; 67:281–4.
Kepley CL, Craig SS, Schwartz LB . Identification and partial characterization of a unique marker for human basophils. J Immunol 1995; 154:6548–55.
McEuen AR, Calafat J, Compton SJ, et al. Mass, charge, and subcellular localization of a unique secretory product identified by the basophil-specific antibody BB1. J Allergy Clin Immunol 2001; 107:842–8.
Mochizuki A, McEuen AR, Buckley MG, Walls AF . The release of basogranulin in response to IgE-dependent and IgE-independent stimuli: validity of basogranulin measurement as an indicator of basophil activation. J Allergy Clin Immunol 2003; 112:102–8.
Dvorak AM . Cell biology of the basophil. Int Rev Cytol 1998; 180:87–236.
Miura K, Schroeder JT, Hubbard WC, MacGlashan DW Jr . Extracellular signal-related kinases regulate leukotriene C4 generation, but not histamine release or IL-4 production from human basophils. J Immunol 1999; 162:4198–206.
Platz IJ, Binder M, Marxer A, et al. Hymenoptera-venom-induced upregulation of the basophil activation marker ecto-nucleotide pyrophosphatase/phosphodiesterase 3 in sensitized individuals. Int Arch Allergy Immunol 2001; 126:335–42.
Binder M, Fierlbeck G, King T, Valent P, Bühring HJ . Individual hymenoptera venom compounds induce upregulation of the basophil activation marker ectonucleotide pyrophosphatase/phosphodiesterase 3 (CD203c) in sensitized patients. Int Arch Allergy Immunol 2002; 129:160–8.
Hauswirth AW, Natter S, Ghannadan M, et al. Recombinant allergens promote expression of CD203c on basophils in sensitized individuals. J Allergy Clin Immunol 2002; 110:102–9.
Boumiza R, Monneret G, Forissier MF, et al. Marked improvement of the basophil activation test by detecting CD203c instead of CD63. Clin Exp Allergy 2003; 33:259–65.
Kahlert H, Cromwell O, Fiebig H . Measurement of basophil-activating capacity of grass pollen allergens, allergoids and hypoallergenic recombinant derivatives by flow cytometry using anti-CD203c. Clin Exp Allergy 2003; 33:1266–72.
Pâris-Köhler A, Demoly P, Persi L, Lebel B, Bousquet J, Arnoux B . In vitro diagnosis of cypress pollen allergy by using cytofluorimetric analysis of basophils (Basotest). J Allergy Clin Immunol 2000; 105:339–45.
Gane P, Pecquet C, Lambin P, et al. Flow cytometric evaluation of human basophils. Cytometry 1993; 14:344–8.
Gane P, Pequet C, Crespeau H, et al. Flow cytometric monitoring of allergen induced basophil activation. Cytometry 1995; 19:361–5.
Knol EF, Mul FPJ, Jansen H, Calafat J, Roos D . Monitoring human basophil activation via CD63 monoclonal antibody 435. J Allergy Clin Immunol 1991; 88:328–38.
Siegmund R, Vogelsang H, Machnik A, Herrmann D . Surface membrane antigen alteration on blood basophils in patients with hymenoptera venom allergy under immunotherapy. J Allergy Clin Immunol 2000; 106:1190–5.
Erdmann SM, Heussen N, Moll-Slodowy S, Merk HF, achs B . CD63 expression on basophils as a tool for the diagnosis of pollen-associated food allergy: sensitivity and specificity. Clin Exp Allergy 2003; 33:607–14.
Gamboa PM, Sanz ML, Caballero MR, et al. Use of CD63 expression as a marker of in vitro basophil activation and leukotriene determination in metamizol allergic patients. Allergy 2003; 58:312–7.
Bühring HJ, Streble A, Valent P . The basophil-specific ectoenzyme E-NPP3 (CD203c) as a marker for cell activation and allergy diagnosis. Int Arch Allergy Immunol 2004; 133:317–29.
Majlesi Y, Samorapoompichit P, Hauswirth AW, et al. Cerivastatin and atorvastatin inhibit IL-3-dependent differentiation and IgE-mediated histamine release in human basophils and downmodulate expression of the basophil-activation antigen CD203c/E-NPP3. J Leukoc Biol 2003; 73:107–17.
Schulman ES, MacGlashan DW, Peters SP, et al. Human lung mast cells: purification and characterization. J Immunol 1982; 129:2662–7.
Valent P, Ashman LK, Hinterberger W, et al. Mast cell typing: Demonstration of a distinct hemopoietic cell type and evidence for immunophenotypic relationship to mononuclear phagocytes. Blood 1989; 73:1778–85.
Look AT, Ashmun RA, Shapiro LH, Peiper SC . Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J Clin Invest 1989; 83:1299–307.
Fukuda M . Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J Biol Chem 1991; 266:21327–30.
Fukuda M, Viitala J, Matteson J, Carlsson SR . Cloning of cDNAs encoding human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. Comparison of their deduced amino acid sequences. J Biol Chem 1988; 263:18920–8.
Carlsson SR, Fukuda M . The lysosomal membrane glycoprotein lamp-1 is transported to lysosomes by two alternative pathways. Arch Biochem Biophys 1992; 296:630–9.
Carlsson SR, Roth J, Piller F, Fukuda M . Isolation and characterization of human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. Major sialoglycoproteins carrying polylactosaminoglycan. J Biol Chem 1988; 263:18911–9.
Watt SM, Bühring HJ, Rappold I, et al. CD164, a novel sialomucin on CD34+ and erythroid subsets, is located on human chromosome 6q21. Blood 1998; 92:849–66.
Zannettino ACW, Bühring HJ, Niutta S, et al. The sialomucin CD164 (MGC-24v) is an adhesive glycoprotein expressed by human hematopoietic progenitors and bone marrow stromal cells that serves as a potent negative regulator of hematopoiesis. Blood 1998; 92:2613–28.
Watt SM, Butler LH, Tavian M, et al. Functionally defined CD164 epitopes are expressed on CD34+: cells throughout ontogeny, but display distinct distribution patterns in adult hematopoietic and non–hematopoietic tissues. Blood 2000; 95:3113–24.
Doyonnas R, Chan JYH, Butler LH, et al. CD164 monoclonal antibodies that block hemopoietic progenitor cell adhesion and proliferation interact with the first mucin domain of the CD164 receptor. J Immunol 2000; 165:840–51.
Nagata K, Hirai H . The second PGD(2) receptor CRTH2: structure, properties, and functions in leukocytes. Prostaglandins Leukot Essent Fatty Acids. 2003; 69:169–77.
Monneret G, Boumiza R, Gravel S, et al. Effects of prostaglandin D(2) and 5-lipoxygenase products on the expression of CD203c and CD11b by basophils. J Pharmacol Exp Ther 2005; 312:627–34.
Azorsa DO, Hyman JA, Hildreth JE . CD63/Pltgp40: a platelet activation antigen identical to the stage-specific, melanoma-associated antigen ME491. Blood 1991; 78:280–4.
Jin-Hua P, Goding JW, Nakamura H, Sano K . Molecular cloning and chromosomal localization of PD-Iâ (PDNP3), a new member of the human phosphodiesterase I genes. Genomics 1997; 45:412–5.
Andoh K, Piao JH, Terashima K, Nakamura H, Sano K . Genomic structure and promotor analysis of the ecto-phosphodiesterase I gene (PDNP3) expressed in glial cells. Biochim Biophys Acta 1999; 1446:213–24.
Goding JW, Grobben B, Slegers H . Physiological and pathophysiological functions of the ecto-nucleotide pyrophosphatase/phosphodiesterase family. Biochim Biophys Acta 2003; 1638:1–19.
Streble A . Durchflusszytometrische Diagnostik der Graspollenallergie unter Einsatz rekombinanter Allergene. Doctoral Thesis at the University of Tübingen 2004.
Grützkau A, Smorodchenko A, Lippert U, et al. LAMP-1 and LAMP-2, but not LAMP-3, are reliable markers for activation-induced secretion of human mast cells. Cytometry 2004; 61A:62–8.
Rubio V, Stuge TB, Sing N, et al. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat Med 2003; 9:1377–82.
Iter G, Malenfant JM, Delabre RM, et al. Increased natural killer cell activity in viremic HIV-1 infection. J Immunol 2004; 173:5305–11.
Chen JW, Murphy TL, Willingham MC, Pastan I, August JT. Identification of two lysosomal membrane glycoproteins. J Biol Chem 1985; 101:85–95.
Acknowledgements
The authors thank Sabrina TREML, Astrid SCHMITT, and Gülay DEMIREL, Tübingen, for flow cytometry and Johannes MADLUNG, Tübingen, for nanoHPLC-ESI-MS/MS analysis. We also thank Dr. Hannes STOCKINGER, Vienna, for confirming the specificity of the WS antibody 80274 for CD13, and Dr. Peter MEIKLE, Dr. Emma PARKINSON, and Prof. Douglas BROOKS, Adelaide, for confirming the specificity of the WS antibody 80280 for CD107a. This work was supported by a grant from the Deutsche Forschungsgemeinschaft, SFB 510-A1 (F.H. and H.-J.B.), by the fortuene-project F1282700 of the university of Tuebingen (H.-J.B) and by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich, SFB grant – project 018/09 (P.V.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
HENNERSDORF, F., FLORIAN, S., JAKOB, A. et al. Identification of CD13, CD107a, and CD164 as novel basophil-activation markers and dissection of two response patterns in time kinetics of IgE-dependent upregulation. Cell Res 15, 325–335 (2005). https://doi.org/10.1038/sj.cr.7290301
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290301
Keywords
This article is cited by
-
The stem cell revolution: on the role of CD164 as a human stem cell marker
npj Regenerative Medicine (2021)
-
Biomarkers in Food Allergy Immunotherapy
Current Allergy and Asthma Reports (2019)
-
Prostaglandin D2 amplifies lupus disease through basophil accumulation in lymphoid organs
Nature Communications (2018)
-
Basophil Activation Test: Old and New Applications in Allergy
Current Allergy and Asthma Reports (2018)
-
The Use of Biomarkers to Predict Aero-Allergen and Food Immunotherapy Responses
Clinical Reviews in Allergy & Immunology (2018)