ABSTRACT
Polar auxin transport plays a divergent role in plant growth and developmental processes including root and embryo development, vascular pattern formation and cell elongation. Recently isolated Arabidopsis pin gene family was believed to encode a component of auxin efflux carrier (Gälweiler et al, 1998). Based on the Arabidopsis pin1 sequence we have isolated a Brassica juncea cDNA (designated Bjpin1), which encoded a 70-kDa putative auxin efflux carrier. Deduced BjPIN1 shared 65% identities at protein level with AtPIN1 and was highly homologous to other putative PIN proteins of Arabidopsis (with highest homology to AtPIN3). Hydrophobic analysis showed similar structures between BjPIN1 and AtPIN proteins. Presence of 6 exons (varying in size between 65 bp and 1229 bp) and 5 introns (sizes between 89 bp and 463 bp) in the genomic fragment was revealed by comparing the genomic and cDNA sequences. Northern blot analysis indicated that Bjpin1 was expressed in most of the tissues tested, with a relatively higher level of transcript in flowers and a lower level in root tissues. Promoter-reporter gene fusion studies further revealed the expression of Bjpin1 in the mature pollen grains, young seeds, root tip, leaf vascular tissue and trace bundle, stem epidermis, cortex and vascular cells. BjPIN1 was localized on the plasma membrane as demonstrated through fusion expression of green fluorescent protein (GFP). Auxin efflux carrier activity was elevated in transgenic Arabidopsis expressing BjPIN1.
Similar content being viewed by others
INTRODUCTION
Charles Darwin had proposed the concept of translocated chemical messengers in higher plants, which finally resulted in the discovery of polar auxin transport in the 1930s1, 2. Studies, including the application of polar auxin transport inhibitors such as TIBA (2, 3, 5-triiodobenzoic acid), have indicated that polar auxin transport plays divergent roles in plant growth and development including apical dominance, tropic growth3, cell elongation, and lateral root development4, 5. Polar auxin transport was also shown to be critical for plant morphogenesis, including leaf vein pattern6, 7, gynoecium morphogenesis8, establishment of bilateral symmetry during embryogenesis9, 10, 11, embryo axis formation12, 13 and etc (for review see 14). Polar auxin transport is an energy-requiring, cell-to-cell process that is specific for indole-3-acetic acid (IAA) and synthetic auxin. The current model of polar auxin transport, chemiosmotic hypothesis, which was independently proposed by Rubery and Sheldrade (1974)15 and Raven (1975)16, posited that polar auxin transport occurs through the action of cellular auxin influx and efflux carriers located in the plasma membrane of transporting cells. It is the basal location of the efflux carrier that is responsible for the polar nature of auxin transport. Jacobs and Gilbert (1983)17 used an immunological approach to demonstrate the basal localization of a putative efflux carrier, which provided the support of the later aspect of the model.
The first molecular support of the model came from the characterization of a candidate auxin influx carrier, AUX1 (auxin resistant1, the Arabidopsis aux1 mutant showed auxin-resistant root growth phenotype and abolished root gravitropic curvature). AUX1 is a presumptive membrane protein similar to amino acid permeases 18 and was specifically expressed in the root. Further proof of AUX1 as an auxin influx carrier was provided by the work of Yamamoto and Yamamoto (1998)19. A breakthrough in the study of polar auxin transport has been marked with the cloning of two genes, Atpin1 and Atpin2, which encoded presumptive auxin efflux carriers20, 21, 22, 23, 24. Pin-formed inflorescence and decreased polar auxin transport activity of the Arabidopsis pin1 mutant indicated that the primary function of the AtPIN1 protein was for polar auxin transport in the inflorescence axis25. The Atpin1 gene, isolated through En-tagging method, was found to encode a protein with similarity to bacterial and eukaryotic carrier proteins. AtPIN1 protein was detected at the basal end of auxin transport-competent cells in vascular tissues 21. These studies strongly suggested that AtPIN1 might act as a transmembrane component of the auxin efflux carrier and play a role in auxin transport from shoot apex to root tissue in stems.
While Atpin1 was found to be widely expressed in all the tissues tested, another member of the pin gene family, Atpin2, showed specific expression in the epidermis and cortex of root, with a similar basal location of the protein in the transport-competent cells23. Thus, the AtPIN2 together with the AUX1 may act as the component of auxin efflux and influx carrier respectively and take part in the auxin transport from the tip to the base of the root (the basipetal stream), which was required for the root gravitropic. The most compelling evidence of AtPIN2 as a component of auxin efflux carrier came from the experiments with AtPIN2-expressing yeast cells which were resistant to toxic fluoroindoles, an IAA analogy22, and released preloaded IAA more rapidly than control cells, suggesting that AtPIN2 transports IAA out of the yeast cells20.
To date several genes encoding auxin efflux carriers, Atpin 1121, Atpin2/eir 122, 23, and Atpin426, have been isolated and some genes encoding PIN-like proteins were detected from Arabidopsis genome. We are interested in the function of auxin and polar auxin transport on plant development. Here we report the cloning, expression pattern analysis and functional characterization of a pin-like gene, Bjpin1, from Brassica juncea through molecular technique and transgenic approaches in Arabidopsis.
MATERIALS AND METHODS
Materials
Enzymes used for DNA restriction and modification were obtained from Boehringer (Mannheim, Germany). DNA primers for polymerase chain reaction (PCR) and "LAPCR in vitro cloning kit" were obtained from TaKaRa Biotechnology (Dalian, China). T3 and T7 mMESSAGE mMACHINETM Kits were obtained from Ambion. Dig-RNA labeling mix, Anti-Pig, were obtained from ROCHE. Western Blue were obtained from Promega. Radiochemicals [α-32P]dCTP and [14C]IAA were obtained from Yahui Company (Beijing, China) and SIGMA (St.Louis, USA) respectively. The leaf l ZAPII cDNA library and genomic library of Brassica juncea were kindly provided by Eng-Chong Pua (National University of Singapore).
Bacteria and plants
Escherichia coli strain XL-1 Blue (Stratagene, La Jolla, USA) was used for library screening and DNA cloning procedures. Agrobacterium strain LBA4404 and GV3101 were used for leaf disc transformation of Nicotiana tobacum cv Gx-1 and floral dip transformation of Arabidopsis thaliana C2427, 28. Brassica juncea and tobacco plants were grown in the green house with a 12 h light (26°C) and 12 h dark (20°C )period. Arabidopsis plants were grown in a phytotron in soil with a 16 h light (22°C )and 8 h dark (15°C) period.
Isolation of the Bjpin1 cDNA and sequence analysis
DNA manipulation was performed using standard protocols29. The nucleotide sequence of Atpin121 and two other Atpin1 homologous sequences (Accession Nos. AC002311, gene T26J12.14, and AC002291, gene F22K20.18, found through searching the GenBank using Atpin1 as bait) were used for designing two reverse degenerated primers pinA1, 5′-AG(A/T/C/G)GG(A/G/T)AT(A/T/- C/G)CCCAT(A/T/C/G)AC-3′ and pinA2, 5′-(A/G)TACCA- (A/G/T)AT(A/G/T)AT(A/G)CA(C/T)TG-3′, corresponding to amino acid VMGIPL and QCIIWY at the position of 115-120 and 140-145 of AtPIN1, respectively. Primers, pinA2 and standard T3, were used for amplification of pin-homologous DNA fragments from the cDNA library of B. juncea. PCR products of about 550bp were subcloned into TA vector and clones were further confirmed by PCR using primers pinA1 and T3. Positive clones were then sequenced. Based on the sequences obtained, specific primers pinS1 (5′-TACGTCGCTATGATCCTCGC-3′) and pinA4 (5′-CGACGACCTGGACCATGAG-3′) were then synthesized for isolation of full-length cDNA from library through PCR-based screening30. Plaque-purified phage clones were converted to pBluescript SK derivative using helper phage Exassist according to supplier's(Stratagene) instructions. The clone pBjpin1, which contained the longest cDNA insert, was used for further analyses.
DNA sequencing was performed by TaKaRa Biotechnology (Dalian, China). Computational analysis was performed with the help of the programs of the Wisconsin Genetics Computer Group (GCG Package, Version 10.1). The BLAST search programs were used for sequence comparisons on DNA and amino acid sequences in GenBank, EMBL, dbEST and SwissProt databases. Sequence alignments were performed using the BESTFIT and PILEUP programs. Hydrophobic analysis was performed with PEPPLOT program.
Isolation of genomic sequence of Bjpin1
Based on the p romoter region of Bjpin1 (see below), specific primers Bjpin103 (5′-TTGACCCAATACACTGTATGG-3′) and Bjpin1S (5′-CTTCTTTCTGGTTGATTAGAGAG-3′) were designed for isolation of Bjpin1 genomic sequences from B juncea genomic library through PCR-based screening. Bjpin1 genomic sequence was amplified with Bjpin103 and standard T3 primers using obtained positive clone as template, and the resulted PCR fragment, which was around 6 Kb, was subcloned into TA vector and sequenced. Sequence alignments were performed using the BESTFIT programs.
Northern blot analysis
Materials were harvested from well-watered B juncea plants. RNA was prepared according to the protocol of Logemann et al. (1987)31. RNA (20 μ per lane) was electrophoretically separated in denaturing 15%(v/v) formaldehyde, 1.5% (w/v) agarose gels and blotted onto Hybond-N+ membranes (Amersham, USA). RNA was fixed onto the membrane by heating for 2 h at 80°C. A 400-bp PCR product of the Bjpin1 (encoded the amino acid residues at position 305-442) was served as [α-32P]dCTP-labeled hybridization probe. Membranes were hybridized at 65°C in 250 mM sodium phosphate buffer (pH 7.2) containing 7% SDS, 1% BSA, and 1mM EDTA. Washes were performed at 65°Cin 2×SSC, 0.5% SDS for 15 min and in 0.2×SSC, 0.5% SDS for 20 min32. Blots were exposed to Fuji X- films between intensifying screens for 4-5 d at −70°C.
Isolation of the Bjpin1 promoter fragment, tobacco transformation and GUS activity assay
“PCR-based DNA walking” technique was employed for isolation of Bjpin1 promoter region. Genomic DNA of B juncea, digested with Hind III, was ligated to the corresponding adaptor according to the supplier' s(TaKaRa, Dalian) instructions. Specific primers BjPS1 (5′-TACAGAGCCGTAGGCGAGGATCATAG-3′), BjPS2 (5′-CGAGGATCATAGCGACGTAGAG-3′) and Bjpin1S (5′-CTTCTTTCTGGTTGATTAGAGAG-3′) were used for PCR amplification. A 320-bp amplified fragment was subcloned into TA vector and then sequenced to confirm the promoter region of Bjpin1 gene. Subsequently, the 320-bp DNA fragment of Bjpin1 promoter region was subcloned into pBI101 binary vector with Hind III/BamH I enzymes. The plasmid, pBjpin1-pro, containing the promoter-uidA coding region, was transformed into A. tumefaciens strain LBA4404 for tobacco transformation. Transformation was performed using leaf disc as materials. GUS activity was determined using 5-bromo-4-chloro-3-indolyl β-D-glucuronic acid (X-gluc) as substrate33.
In situ hybridization
In situ hybridizationof fixed and paraffin embedded flowers of B juncea was performed as described34. PCR fragment of Bjpin1 (position 1041-1451) were ligated into pBSK vector through Sma I cleavage site generating pSKpinV for Bjpin1-specific RNA probes. After pSKpinV linearizing BamH I for antisense and Hind III for sense transcription in vitro transcription and digoxigenin labeling were performed, signals were developed with 100 μl Western Blue per slide and observed under dark field (Olympus microscope).
Bjpin1-GFP fusion studies
The 2 kb Bjpin1 coding region (just before the stop coden), which was amplified with primers T3 and Gbjpin1 (5′-GGGGTACCTAACCCGAGTAGAATATAG-3 , Kpn I site italic), was cut with Kpn I and Xba I and then ligated to the precut (with same restriction enzymes) binary vector p12G, which contained CaMV35S promoter and GFP (green fluorescent protein) coding region (B Zhang, Shanghai Institute of Plant Physiology, Shanghai). Resulted construct, p12G-Bjpin1, was sequenced to confirm that no nucleotide substitutions occurred during the PCR amplification and in frame of the GFP coding region. The p12G-Bjpin1 was then transformed into A. tumefaciens strain LBA4404 for tobacco leaf disc transformation. Resistant tobacco plants were confirmed to be transgenic plants via Northern blot analyses using GFP coding region as hybridization probe.
Roots of transgenic tobacco were transferred onto microscope cover slips and imaged on a fluorescence microscope equipped with a UV lamp and filter (Zeiss GFP filter, =488nm, λex=507 nm). Plasmolysation of root cell was performed with 20% sucrose solution treatment.
Assay of IAA transport activity in transgenic Arabidopsis inflorescence axes
The 2.6 kb DNA fragment, which was cut with EcoR I from plasmid pBjpin1 and included the full Bjpin1 gene, was blunted with T4 polymerase. Resulted DNA fragments were then subcloned to the Sma I site of the binary vector pBinAR-kam (from Max-Plank-Institute of Molecular Plant Physiology, Germany) to generate the construct harboring Bjpin1 in both forward and reverse orientations. Constructs with different orientations were checked with restriction enzymes and then transformed into A. tumefaciens strain GV3101. Transgenic Arabidopsis plants were obtained using floral dip method and confirmed by PCR using primers spanning the CaMV35S and Bjpin1 coding region, with genomic DNA as templates.
IAA polar transport activity assay was performed as described by Okada et al. (1991). In short, 2.5 cms of inflorescence axes (at same position in different plants) of transgenic arabidopsis plants were cut and incubated in 1/2 MS35 solution containing 1.45 μ M [14C]IAA in both normal and inverted orientations for 18 or 24 h. After 2 d [14C] radioactivity was measured by liquid scintillation counting (Beckman LS650) with 3 ml counting solutions. Measurement were performed with: 1) 0.5 cm ends of inflorescence axes; 2) both halves (1.25 cm) of inflorescence axes after incubation.
RESULTS
Cloning of Bjpin1 from B juncea
Sequences analyses of a DNA fragment of around 550 bp, amplified from B juncea cDNA library with T3 and degenerated primers revealed that it encoded a peptide sharing 75% identities with AtPIN1 at positions 1 ∼ 145, which indicated a putative PIN-like fragment of B juncea. A full-length cDNA, designated as Bjpin1, encoding a PIN-like protein was then isolated from B juncea. The Bjpin1 cDNA has a length of 2557 nucleotides with an open reading frame present between nucleotides 126 (ATG) and 2042 (TGA). The predicted protein contained 639 amino acid residues and had a molecular mass of ∼ 70 kDa, similar to that of previously reported AtPIN121 and AtPIN2/EIR122. The calculated isoelectric point of BjPIN1 was 8.88.
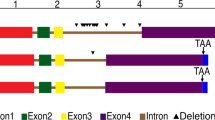
A genomic fragment harbouring the entire Bjpin1 coding region was isolated through library screening. A comparison between the genomic DNA and cDNA sequences revealed the presence of 6 exons (varying in size between 65 bp and 1229 bp) and 5 introns (sizes between 89 bp and 463 bp) in the genomic fragment (Fig 1A). The complete Bjpin1 gene had a size of approximately 5 kb, including 5′- and 3′- untranslated regions, and putative 5′ regulatory elements.
Gene organization, sequence alignment and hydrophobic analyses of BjPIN1 A. Exon / intron structure of the Bjpin1 gene. Exons are presented as boxes. Numbers indicate exon and intron sizes (nucleotides). B. Alignment of BjPIN1 with other PIN proteins. Numbers on the top indicate amino acid positions. Accession numbers of cDNAs were as follows: AtPIN1, AF089084; AtPIN2, AF086906; AtPIN3, AF087818; AtPIN4, AF087016; AtPIN6, AF087819; AtPIN7, AF087820. C. Hydrophathy analysis of BjPIN1, 11 putative transmembrane regions were marked.
Structural organization of the BjPIN1 protein
Fig 1B shows the alignment of BjPIN1, AtPIN1, AtPIN2 and other PIN-like proteins in Arabidopsis. BjPIN1 shared high homology with Arabidopsis PIN proteins (Tab 1), i.e. 65.1% and 64.4% identities with AtPIN1 and AtPIN2, respectively, and the highest identities (92.7% and 87%, respectively) were found with the putative AtPIN3 and AtPIN7 (accession No. AF087818 and No.AF087820, respectively). Conserved regions were found at N- and C-termini of all the proteins. Hydropathy analysis showed BjPIN1 contained highly hydrophobic domains in both N- and C-terminus of proteins, which are conserved in all PIN and PIN-like proteins (Fig 1B), and a varied central region that is predominantly hydrophilic (Tab 2). Structural analysis indicated the presence of around 11 predicted transmembrane regions in BjPIN1 (Fig 1C), which was similar with AtPIN1.
Expression pattern analyses of Bjpin1
The cDNA fragment of Bjpin1 encoding the varied region (amino acid residues position 305 ∼ 442) was used as a hybridization probe to detect Bjpin1 transcription levels. As shown in Fig 2 a single band of transcripts of approximately 2.5 kb was detected, consistent with the length of the Bjpin1 cDNA. The Northern blot revealed the presence of Bjpin1 mRNA in various B juncea tissues, including hypocotyls, cotyledons, leaves, stem, flowers and fruits, with a relatively higher and lower level of expression in floral and root tissues.
To further study the expression pattern of Bjpin1, a 316-bp promoter region of Bjpin1 was isolated by "genome walking"and was fused to the E. coli β-glucuronidase (uidA) coding sequence. The construct was stably integrated into the tobacco genome via Agrobacterium-mediated transformation and confirmed via PCR amplification using the primers of CaMV35S promoter and GUS coding gene (Fig 3A). From 10 independent transgenic tobacco lines analyzed, 3 lines showed clear GUS activities in root tip (Fig 3B) leaf vein (Fig 3C) young seeds (Fig 3D) mature anther and pollen grains (Fig 3E) and stem. In stem, GUS activities were detected in epidermis, cortex and vascular cells, and particularly, the leaf trace cells (data not shown). Further in situ hybridization experiment of flowers confirmed the expression of Bjpin1 in the pollen grains (Fig 3F sense control not shown).
Expression of Bjpin 1 in various tissues A. Confirmation of transgenic plants containing construct harbouring promoter-gus regions. PCR amplification was performed with primers from promoter and gus coding region using genomic DNA as template. From left to right: 1: 1Kb ladder; 2: using plasmid as template; 3: using genomic DNA from untransformed plants; 4–6: using genomic DNA of transformed plants. B-E: GUS activities were detected in root tip (B), leaf vein (C), seeds (D) and pollen grain (E). F: In situ hybridization using B juncea anther as material. Arrows showed the expression of Bjpin1 in pollens.
BjPIN1 was localized on plasma membrane
Based on the chemiosmotic hypothesis, auxin efflux carrier was located on plasma membrane for function of polar auxin transport. To study the cellular localization of BjPIN1, the CaMV35S-Bjpin1-gfp construct resulting in the expression of Bjpin1-GFP fusion proteins, was transferred to tobacco plants. Of the 5 resistant tobacco lines, two of them showed high gfp mRNA levels according to the Northern blot using gfp coding sequence as the hybridization probe (Fig 4A). Image by fluorescence microscopy showed much stronger fluorescencein roots of the transgenic plants (Fig 4B) and the fluorescence was detected only on the surface of cell (Fig 4C) with the polar localization in root tissues(Fig 4D). Root cells of transgenic tobacco were then plasmolysed and fluorescences were found to be restricted to the plasma membrane of plasmolysed cells (Fig 4E, F). Thus the BjPIN1-GFP fusion studies suggested BjPIN1 maybe a plasma membrane protein with the polar localization, which is consist with its function as an auxin efflux carrier.
Cellular localization of BjPIN1 through GFP fusion studies A. Northern blot analysis of the transgenic tobacco plants using gfp coding region as probe. Two lines showed positive signals. B.Roots under UV fluorescences (upper-control; bottom-transgenic plants); C-D.Fluorescences were localized on the surface of root cells (C) with polar localization (D); E-F. Plasmolysed root cells of transgenic tobacco, under normal light (E) and UV fluorescences (F).
Polar auxin transport activity in transgenic Arabidopsis
To decide whether Bjpin1 encodes a functional auxin efflux carrier, IAA transport assay was performed on transgenic Arabidopsis plants. Both binary vectors harboring forward and reverse orientations of Bjpin1 under a CaMV35S promoter were used to transform Arabidopsis. Measurements of polar auxin transport activity in the inflorescence axes with [14C]IAA were performed with either 0.5 cm ends of inflorescence axes or both halves (1.25 cm) of inflorescence axes after incubation (to compare the ratio of [14C] in both halves) and the results showed that most of the transgenic plants overexpressing BjPIN1 possessed an increased auxin transport activities in comparison with control plants. As shown in Tab 3, most of the transgenic plants tested showed increased auxin transport activities, two transgenic lines, T-013 and T-016, had around 2 times higher auxin transport activities than the control. The results, to a certain extent, demonstrated that BjPIN1 was a functional auxin efflux carrier. However, measurement of inflorescence axes of transgenic plants harbouring antisense orientation of Bjpin1 did not show decreased polar auxin transport activities.
DISCUSSION
Researches on the function, expression pattern and location of recently isolated auxin carriers18, 21, 22, 23, 26 have provided a great wealth of date for understanding the physiological role of auxin and polar auxin transport during plant development. Till now, around 8 different pin-like genes have been found in Arabidopsis genome and a few homologous genes from rice as well. Two of the Arabidopsis genes, Atpin1 and Atpin2, have been investigated in detail21, 23. Since auxin transport plays divergent roles in plant development, it is possible that some of the pin genes share functional redundancy and every individual member of the PIN family has its distinct functions. It is thought that there are tissue- or even cell-specific auxin efflux carriers that are responsible for multiple auxin transport pathways36, as indicated by the distinct function of AtPIN1 and AtPIN2/EIR1/AGR1 in the shoot and root, respectively21, 22, and by predominantly lateral localization of AtPIN3 in endodermal cells37. The Arabidopsis mutant pin1 shows normal tropic responses of shoot and root25 also indicated that there maybe other pin genes that controlled the lateral auxin transport pathways, according to the Cholodny-Went hypothesis38. Models for lateral auxin transport pathway present in shade avoidance have been proposed39. There are at least 5 pin-like genes present in B juncea and all of them show different expression patterns40, which indicates that there is also a pin-gene family in B. juncea genome and different members probably play different roles. Distribution of auxin is critical for plant development, especially for plant pattern formation, and auxin efflux carriers are responsible for auxin distribution in plant tissues (cells). Isolation and expression pattern analysis of other pin-like genes will provide informative clues to the role of each PIN protein in plant growth and development.
BjPIN1 has a similar size (70 kDa) with AtPIN1 and other PIN-like proteins and share high identities with them (highest identity with AtPIN3). They all have similar structural organizations, i.e. conserved, highly hydrophobic domains in both N- and C-terminus and a varied domain present in the central region that is predominantly hydrophilic. However, several genomic sequences, found through database searching, encoding putative PIN-like proteins are relatively unconserved at N- or C- termini. It is still unclear whether these were due to the annotation of genomic structure or the presence of PIN-proteins with different structural organizations.
Northern blots indicated that the Bjpin1 was expressed in most of the tissues tested with a relatively high level of expression in flowers, however, promoter-GUS fusion studies in transgenic tobacco further revealed the expression of Bjpin1 in root tips, stem epidermis, cortex, vascular cells, and particularly the leaf trace cells. This is still unclear as this might due to the short promoter region used or the expression in heterologous plant species. Anyway, the expression pattern of Bjpin1 was different from that of Atpin1 that was expressed variety in all tissues, while Atpin2 was specifically expressed in root tissues. Promoter-GUS fusion studies revealed a strong expression of Bjpin1 in mature pollen grains, which was consistent with the results of Northern blot analyses, and the pollen expression was further confirmed by in situ hybridization. It is important to note that two pollen-specific cis-elements, “AGAAA” are found in Bjpin1 promoter region (both putative"TATA"and "CAAT"cassettes were detected). However, a longer promoter region of Bjpin1 is needed for detailed expression pattern analyses. All these, together with the expression of Bjpin1 in epidermis and cortex of stem, suggested that BjPin1 may be involved in flower, particularly pollen grain development, and differential growth in stem through directly auxin transport.
According to chemiosmotic hypothesis41 it was the basal location of efflux carriers on the plasma membrane of transporting cells that were responsible for the polar nature of auxin transport. This has been confirmed by the polar localization of AtPIN1 and 2. Here, through GFP fusion studies, we showed BjPIN1 was a plasma membrane protein, which was consistent with its function as a putative auxin efflux carrier.
Polar auxin transport assay of transgenic Arabidopsis plants with increased level of BjPIN1 showed higher polar auxin transport activities than control plants. It has been reported that deficiency of Atpin1 resulted in the decrease of polar auxin transport in the inflorescence axis of Arabidopsis25. However, antisense approach of Bjpin1 in Arabidopsis did not show an obvious decrease of auxin transport in inflorescence axis, this may be due to the relatively low homology of Bjpin1 to Atpin1. To study the physiological role of Bjpin1 it is necessary to have the transgenic B juncea for detailed analysis.
**The nucleotide sequences reported in this paper have been submitted to the GenBank™/EMBL Data Bank with accession numbers AJ132363 (Bjpin1 cDNA), AJ307883 (promoter region of Bjpin1) and AJ307884 (genomic sequence of Bjpin1).
References
Darwin C . Power of movement in plants. 1880; John Murray, London.
Lomax TL, Muday GK, Rubery PH . Auxin transport. In PJ Davies, ed, Plant hormones. Ed2. 1995; Kluwer Academic Publishers, Dordrecht, The Netherlands, pp509–530.
Evans ML . Gravitropism: interaction of sensitivity modulation and effector redistribution. Plant Physiol 1991; 95:1–5.
Reed RC, Brady SR, Muday GK . Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 1998; 118:1369–78.
Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 2001; 13:843–52.
Mattsson J, Sung ZR, Berleth T . Response of plant vascular systems to auxin transport inhibition. Development 1999; 126:2979–91.
Sieburth LE . Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol 1999; 121:1179–90.
Nemhauser JL, Feldman LJ, Zambryski PC . Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000; 127:3877–88.
Liu CM, Xu ZH, Chua NH . Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 1993; 5:621–30.
Fischer-Iglesias C, Sundberg B, Neuhaus G, Jones AM . Auxin distribution and transport during embryuonic pattern formation in wheat. The Plant J 2001; 26:115–29.
Hadfi K, Speth V, Neuhaus G . Auxin-induced developmental patterns in Brassica juncea embryos. Development 1998; 125:879–87.
Przemeck GKH, Mattsson H, Hardtke CS, Sung ZR, Berleth J . Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 1996; 200:229–37.
Hardtke CS, Berleth T . The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. The EMBO J 1998; 17:1405–11.
Berleth T, Sachs T . Plant morphogenesis: long-distance coordination and local patterning. Current opinion in Plant Biology 2001; 4:57–62.
Rubery PH, Sheldrake AR . Carrier-mediated auxin transport. Planta 1974; 118:101–21.
Raven JA . Transport of indole acetic acid in plant cell in relation to PH and electrical potential gradients, and its significance for polar IAA transport. New Phytol 1975; 74:163–72.
Jacobs M, Gilbert SF . Basal localization of the presumptive auxin transport carrier in pea stem cells. Science 1983; 220:1297–300.
Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, et al. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Nature 1996; 273:948–50.
Yamamoto M, Yamamoto KT . Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2, 4-dichlorophenoxyacetic acid on the gravitropic response of roots on an auxin-resistant mutant of arabidopsis, aux1. Plant Cell Physiol 1998; 39:660–4.
Chen R, Hilson P, Sedbrook J, Rosen E, Casper T, Masson PH . The Arabidopsis thaliana AGRAVITROPIC1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Nat Acad Sci USA 1998; 95:15112–7.
Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K . Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998; 282:2226–30.
Luschnig C, Gaxiola R, Grisafi P, Fink GR . EIR1, a root specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 1998; 12:2175–87.
Müller A, Guan C, Gälweiler L, Tanzler P, Huijser P, Marchant A, et al. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 1998; 17:6903–11.
Utsuno K, Shikanai T, Yamada Y, Hashimoto T . AGR, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane protein family member. Plant Cell Physiol 1998; 39:1111–8.
Okada K, Ueda H, Komaki MK, Bell CJ, Shimura Y . Requirement of the auxin polar transport in the early stage of Arabidopsis floral bud formation. Plant Cell 1991; 3:677–84.
Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, et al.. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 2002; 108:66–73.
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT . A simple and general method for transferring genes into plants. Science 1985; 227:1229–31.
Clough SJ, Bent AF . Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant J 1998; 16:735–43.
Sambrook J, Fritsch EF, Maniatis T . Molecular cloning: A laboratory manual, Ed2, 1989; Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Alfandari D, Darribere T . A simple PCR method for screening cDNA libraries. PCR Methods and Applications 1994; 4:46–9.
Logemann J, Schell J, Willmitzer L . Improved method for the isolation of RNA from plant tissues. Anal Biochem 1987; 163:21–6.
Fieuw S, Müller-Räber B, Galvez S, Willmitzer L . Cloning and expression analysis of the cytosolic NADP+-dependent isocitrate dehydrogenase from potato: implications for nitrogen metabolism. Plant Physiol 1995; 107:905–13.
Jefferson RA, Kavanagh TA, Bevan MW . GUS fusions: b-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 1987; 6:3901–7.
Jackson D . In situ hybridization in plants. In SJ Gurr, MJ Mcpherson, DJ Bowles, eds, Molecular plant pathology: a practical approach. V1. 1992; Oxford University Press, New York. Pp163–174.
Murashige T, Skoog F . A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 1962; 15:473–97.
Jones AM . Auxin transport: down and out and up again. Science 1998; 282:2201–02.
Bisseling T, Weigel D . Plant development: from cell fate to organ formation (meeting report). Plant Cell 2001; 13: 222.
Cosgrove DJ . Gravitropism of cucumber hypocotyls: biophysical mechanism of altered growth. Plant Cell Environ 1990; 13:235–41.
Morelli G, Ruberti I . Shade avoidance responses. Driving auxin along lateral routes. Plant Physiol 2000; 122:621–26.
Ni WM, Chen XY, Xu ZH, Xue HW . A Pin gene families encoding components of auxin efflus carriers in Brassicajunce. Cell Research 2002; 12(3):247–55.
Goldsmith MHM . The polar transport of auxin. Annu Rev Plant Physiol 1977; 28:439–78.
Acknowledgements
Studies were supported bythe “National Natural Science Foundation of China, No. 30070073” State Key Project of Basic Research, No. G1999011604" and “National Natural Science Foundation of Pan-Deng”. We thank Dr. Charles Brearley and Jian Xu for helpful discussions on the manuscript. We greatly thank Dr. Klaus Palme for providing the Atpin1 nucleotide sequences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
NI, W., CHEN, X., XU, Z. et al. Isolation and functional analysis of a Brassica juncea gene encoding a component of auxin efflux carrier. Cell Res 12, 235–245 (2002). https://doi.org/10.1038/sj.cr.7290130
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290130
Keywords
This article is cited by
-
Pseudo almost periodic solutions and global exponential stability of a generalized population model with delays and harvesting term
Computational and Applied Mathematics (2022)
-
Gravity-controlled asymmetrical transport of auxin regulates a gravitropic response in the early growth stage of etiolated pea (Pisum sativum) epicotyls: studies using simulated microgravity conditions on a three-dimensional clinostat and using an agravitropic mutant, ageotropum
Journal of Plant Research (2007)
-
A Pin gene families encoding components of auxin efflux carriers in Brassica juncea
Cell Research (2002)